ImageJ for FCS Express Knowledge Base
Workflow for analyzing ImageJ data in FCS Express:
The flowchart below indicates how the DNSMacroExample.ijm macro can be used to export ImageJ data as .ijout files readable by FCS Express (Fig. T28.116). The DNS-IJXwriter.xlsm macro (written by Peter Haub) can be used to define the relationship between grouped images.
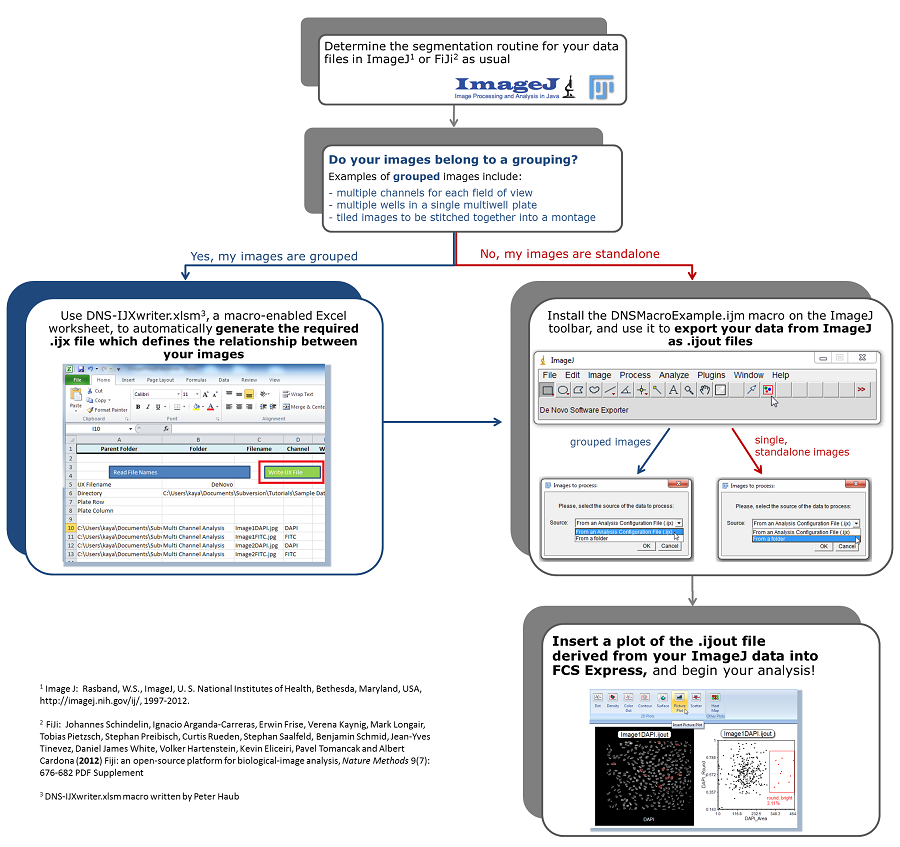
Figure T28.116 Workflow for analyzing single or grouped images segmented in ImageJ within FCS Express
ImageJ Macros for use with FCS Express can be downloaded from http://www.denovosoftware.com/site/imageJ.shtml.
Macros are installed in ImageJ with the following steps:
| 1. | Open ImageJ |
| 2. | Click on Plugins→Macros→Install or press control+shift+M |
| 3. | Navigate to the location of the macro |
| 4. | Click Open. |
The macro will now be installed in the ImageJ tool bar. For more help with ImageJ Macros please see the ImageJ website at http://rsbweb.nih.gov/ij/index.html.
Folder Structure and File Naming:
File Names:
The DNSExampleMacro will name any exported file names (.ijout files) and folder names with the name of the original image. For instance, if your original image was named DapiImage#1.tif, the resulting ImageJ export file will be named DapiImage#1.ijout and a folder will be created name DapiImage#1 that contains a .results.txt file named DapiImage#1.results.txt and a mask image named DapiImage#1.mask.tif. Note that the name of the .ijout file and the folder that contains the results must always be the same.
Loading Files (Folder Structure):
There are options in FCS Express User Options→Data Loading→ImageJ Options for choosing how to load your data (figure T28.117).
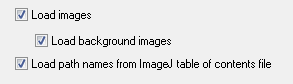
Figure T28.117 The ImageJ Data Loading options available from Edit Preferences in FCS Express 4 Image Cytometry.
Choosing Load images or Load background images will determine if you want to load just raw data (Load images unchecked), or raw data plus the original images (Load images checked), and/or background masking (Load background images checked to load masks/background or unchecked to not load masks/background).
"Load path names from ImageJ table of contents file" indicates that you would like to load the data files with the absolute file path that was determined at the time of exporting the data from ImageJ. If you have subsequently moved the data files or folder containing the data you will need to uncheck this button. To work with data without loading the path names from the ImageJ table of contents file, the following files must be within the same folder at the same level as in figure T28.118 below:
| • | The original image (DNAImage1.jpg below) |
| • | The exported .ijout file (DNAImage1.ijout below) |
| • | The folder containing the .results.txt file and the .mask.tif file (DNAImage1 folder below) |
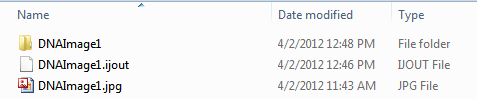
Figure T28.118 When moving .ijout files to another location on your computer the .ijout file, original image file, and folder containing the .results.txt and .masks.tif files must be at the same level.
Note that the name of the .ijout file and the folder that contains the results must always be the same. Multiple segmentations, and therefore multiple .ijout and results folders may also be generated from the same images. In the case of running multiple segmentations the same set of original images may be used without duplicating the image data set.
Working with Plate Based Data:
If the .ijx file represents a plate, the lines of the file that are related to a well must be grouped consecutively. For example, if the file is about a plate with wells D05 and H04, all the channels for well D05 must be in a group of lines, followed by another group of lines with the channels of well H04.
If the ijx file represents a plate, the last line must have the plate information, starting at column 1 with a # and the words 'Plate Info'. Then it is expected a field with the constant 'Rows', the number of rows in the plate, the constant field 'Columns', and the number of columns of the plate. For example: #Plate Info,Rows,8,Columns,12 . See figure Figure T28.119 for an example.
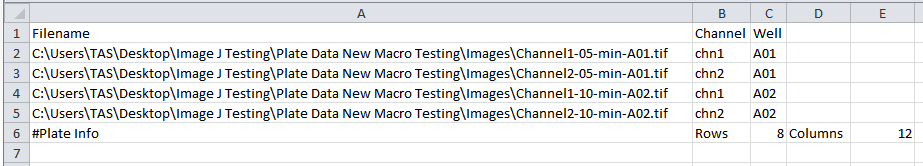
Figure T28.119 An .IJX Analysis Configuration file for a plate based assay that contains two wells, one image per well, and each image contains two channels. The text #Plate Info, Rows, 8, Columns, 12 indicate the plate is 8 x 12 rows, or a 96 well plate.
The name of the .ijx schema can be customized. At the bottom of the DnsAnalysisCreator.ijm you will find:
// This function returns the name of the automatically generated .ijx file.
function getExporterFilename() {
return "FileListForDnsExporter";
Modify the text in, 'return "XXXX;' where the X's represent your desired filename, to change to a customized file name.
If this macro file is installed in ImageJ (Plugins→Macros→Install), a blue rectangle with the hint "De Novo Software Analysis Configuration File Creator" will be shown in the toolbar (figure T28.120).
Figure T28.120 The ImageJ tool bar with the DnsAnalysisCreator.ijm macro installed. It is represented by a black box with the hint "De Novo Software Analysis Configuration File Creator".
