DNA Cycle and Model Summary Reference
MultiCycle AV for FCS Express is capable of autofitting the appropriate Cycle to suit your data. However, users have the ability to override the autofit and choose a Cycle to fit to their data.
Here is a summary of the various Cycles that are available in MultiCycle AV.
1 Cycle |
The most common and frequently used. Used when there is 1 full cell cycle present. |
1 Cycle Standard Overlay Peak |
Similar to 1 Cycle. Ideal when using a standardization tool like chicken erythrocytes. |
1 Cycle Apoptotic Overlay Peak |
This cycle is performed when it is assumed that there is one extra peak to the left of the G0/G1 peak. |
1 Cycle Aneuploid Overlay Peak |
This cycle is performed when there is an Aneuploid peak present that does not contain a full cell cycle. |
1 Cycle Diploid Overlay Peak |
This cycle is performed when there is a Diploid peak that does not contain a full cell cycle. |
2 Cycle |
This cycle is used when there are 2 full cell cycles present. |
3 Cycle |
This cycle is used when there are 3 full cell cycles present. |
Synchronous S Phase |
This cycle should be used when performing an experiment where S phase is arrested, leading to a peak in S peak, rather than a smooth gradual curve. |
MultiCycle AV for FCS Express applies six different models to your data at the same time.
In FCS Express, you can create a DNA Model Statistics Summary box that shows your statistics as it applies to each of the six different models (Figure 25.15).
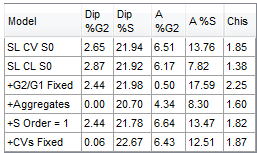
Figure 25.15 - DNA Model Statistics
Here is a summary of the various Cycles that are available in MultiCycle AV.
SL S0 |
Most common model used. Sliced nuclei background modeling with zero order S phase, but without other options. Used when debris curve looks like a ski slope. |
SL CL S0 |
Sliced nuclei background modeling with clumping compensation with zero order S phase. Used if debris curve is flat and cuts into G0/G1. |
+G2/G1 Fixed |
Sliced nuclei background modeling, plus the constraint of the G2/G1 ratio. For 1 cycle fitting models, the G2/G1 ratio is the current default stored within Multicycle AV.
Please note, for 2 Cycle models, the G2/G1 ratio of the diploid cycle is constrained to be equal to the aneuploid G2/G1 ratio that resulted from the user-chosen fitting model.
For 3 Cycle models, the G2/G1 ratio of all 3 cell cycles is constrained to be equal to the average of the 2 aneuploid G2/G1 ratios that resulted from the user-chosen fitting model. |
+Aggregates |
Sliced nuclei modeling plus aggregation modeling and addition of G2/G1 constraint, as detailed above. Basically, this is a software version of doublet discrimination. This model looks at all possible combinations of G1 and G2 stuck together and figures out the % of cells that are stuck together. |
+S Order = 1 |
A 1st order S phase polynomial is used with sliced nuclei background modeling. This model is to be used when the S phase is more in the shape of a broadened trapezoid.
S order = 0 (broadened rectangle) S order = 1 (broadened trapezoid) S order = 2 (higher on the sides, lower in the middle) |
+CVs Fixed |
Sliced nuclei modeling with 1st order S phase, plus the CVs of all peaks are constrained. This model can be used when the G2/M peak is buried in the S phase. |
References for this information can be found at our Multicycle Knowledgebase Page.
In cases where the data is suboptimal or the cell cycle peaks are not well defined, users can get Intra-model or Inter-model errors. You can find the definition for these errors at http://www.denovosoftware.com/site/kb-DNAMulticycleInfo.shtml.
