Selecting Individual Parameters for CellProfiler Export
In this section, you will learn:
•How to export a selected number of parameters of interest from CellProfiler rather than all of the measured values.
In the previous export sections we have chosen to export all of the object size, shape, and intensity parameters that are generated by CellProfiler. While some of these parameters may be useful for your experiment not every parameter defined by CellProfiler is useful for every experiment. In order to specifically choose which parameters you would like to export you must split the export process into two modules: one for the Image.CPTOC and one for the selected parameters in the .CPOUT files. This tutorial will guide you through the process by modifying the Section5pipeline.cpproj pipeline located in the Tutorial Sample Data archive. The pipeline as been designed for use with the Exporting Single Images tutorial which should be completed before beginning this tutorial.
1. Load the Section5pipeline.cpproj pipeline in CellProfiler from the FCS Express Tutorial Sample Data folder.
2.Confirm the default input and output folders in CellProfiler to load and store the data as shown in Figure T24.13.
3.Right click on ExportToSpreadsheet module.
4.Choose Duplicate from the dialog.
5.Click on the first ExportToSpreadsheet module to select it.
6.Click on the Remove this data set button in the Data to export Nuclei (nuclei_time.cpout) data set.
7.Click on the Remove this data set button in the Data to export Cells (cells_time.cpout) data set.
(Note: Only the Data to export Image (Image_time.cptoc) dataset will be remaining in this module as in Figure T24.39)
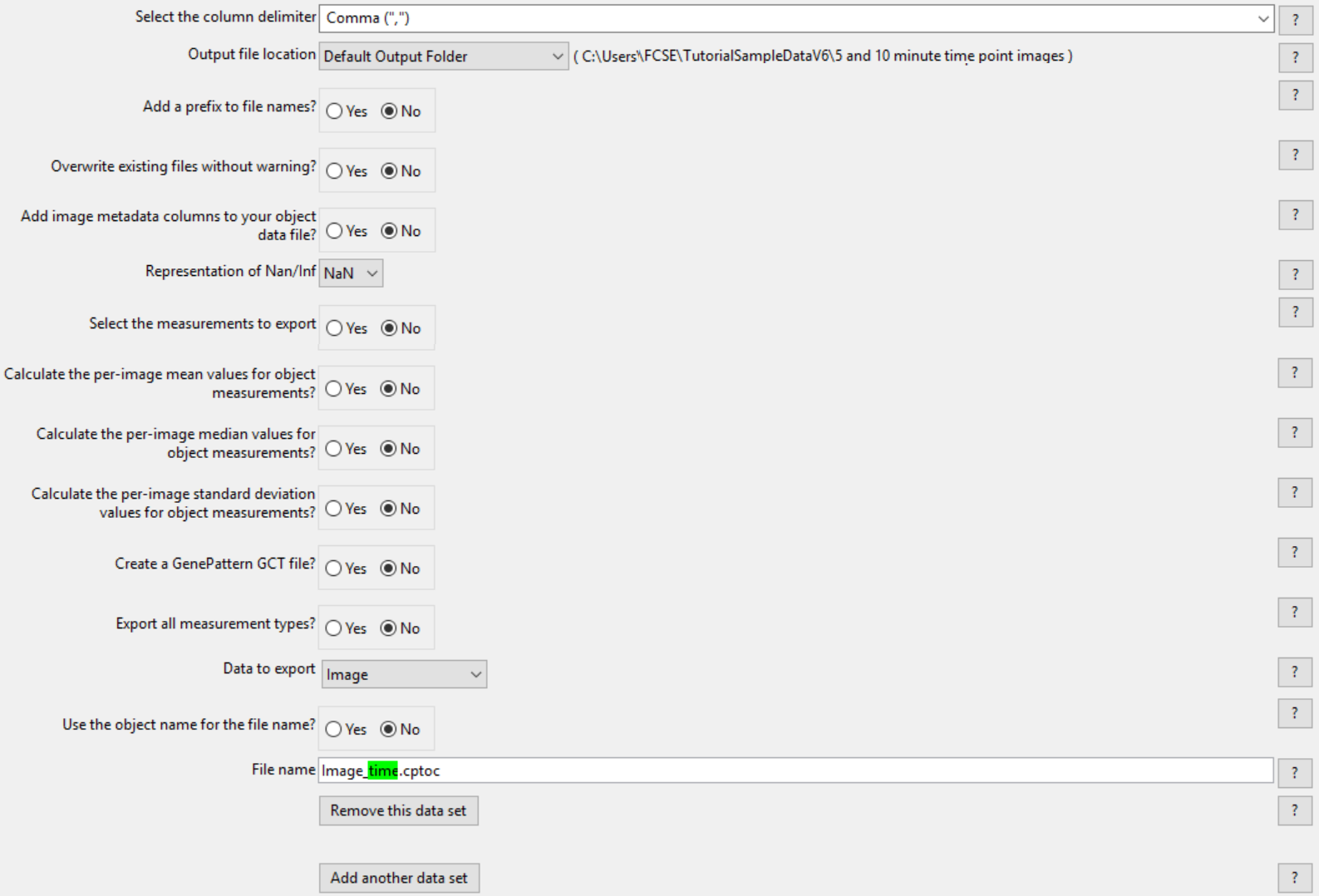
Figure T24.39 The first ExportToSpreadsheet module after the Data to export Nuceli and Cells has been removed.
8. Click on the second ExportToSpreadsheet module to select it.
9. Click on the Remove this data set button in the Data to export Image (Image_time.cpout) data set.
(Note: Only the Data to export Nuclei (nuclei_time.cpout) data set and Data to export Cells (cells_time.cpout) data set will be remaining in this module as in figure T24.40)
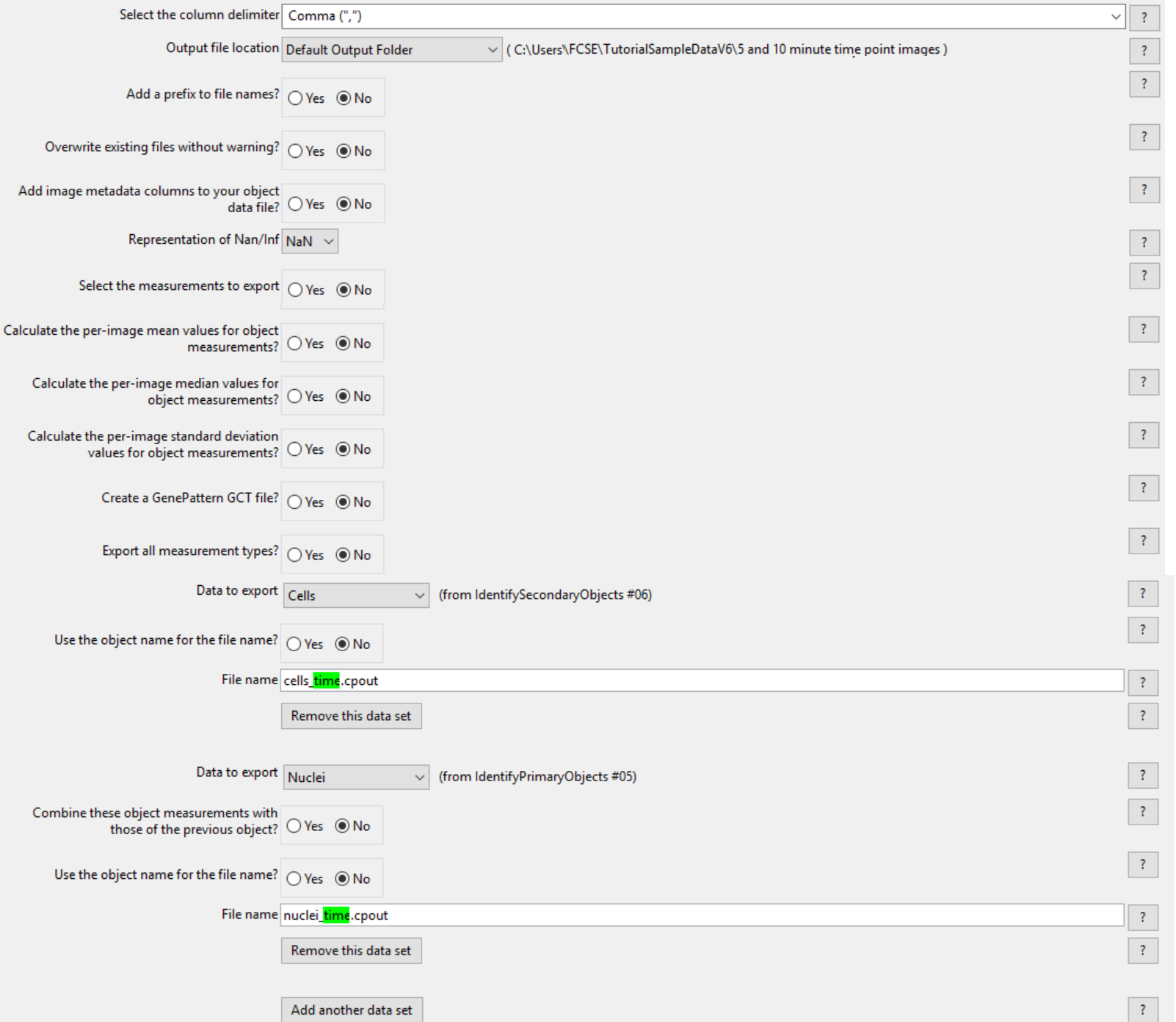
Figure T24.40 The second ExportToSpreadsheet module after the Data to export Image has been removed.
10. Click Yes radio button in the Select the measurements to export (Figure T24.41).
11. Click the Press button to select measurements (Figure T24.41).
12. Expand the Cells→AreaShape categories (Figure T24.41).
13. Check the box for Area (Figure T24.41).
14. Check the box for the Cells→Intensity→MeanIntensity category (Figure T24.41).
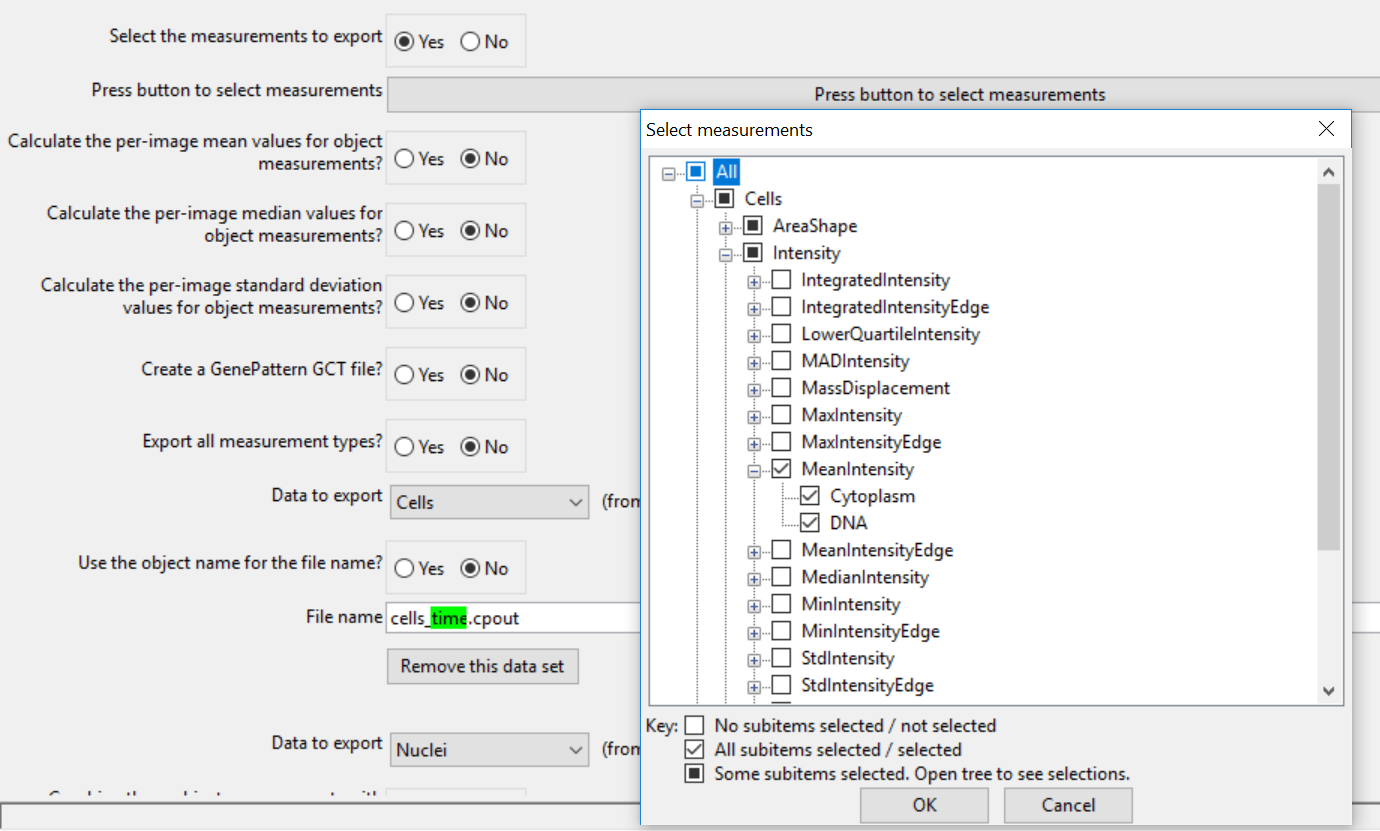
Figure T24.41 Choosing the Select the columns of measurements to export check box, clicking on the Press to select measurements button, and selecting the parameters to export for the Cells objects.
15. Expand the Nuclei→AreaShape categories
16. Check the box for Area.
17. Expand the Nuclei→Intensity→MeanIntensity category.
18. Check the box for Cytoplasm.
19. Check the box for DNA.
20. Click OK.
21. Click Analyze images to run the pipeline.
The ExportToSpreadsheet module has now been broken down into a module for the Image.cptoc and a module for the cells.cpout and nuclei.cpout. The Image.cptoc will use the default export process to export all the information FCS Express needs to import your data. The .cpout files will now only contain the parameters for AreaShape_Area, Intensity_MeanIntensity_Cytoplasm, and Intensity_MeanIntensity_DNA. After analysis is complete you may open the cells.cpout and nuclei.cpout files in FCS Express to view the new data set as you did in the Importing Single Image Data section.
