Exporting Large Tiled Images From CellProfiler
Exporting Large Tiled Images
Often high resolution imaging of large objects or specimens is accomplished by tiling many smaller images to create one large high resolution image (also referred to as a montage image). These images are usually provided from the acquisition instrument or software in one of two ways. Often, they are automatically stitched together and provided as one large image. In this case, they must be broken into smaller images for CellProfiler to analyze (due to memory limitations of CellProfiler) and then FCS Express will reassemble the larger image. Alternatively, they are provided as many smaller images. In that case, CellProfiler can analyze the smaller images as they are, and FCS Express will (optionally) reassemble them into a single large image.
In the course of this tutorial we will explain:
•Image naming conventions needed for FCS Express to import the tiled data processed by CellProfiler.
•How to split large images into tiles for segmentation in CellProfiler.
•Exporting tiled images in CellProfiler for use in FCS Express
Naming Conventions for Tiled Images
When reconstructing a series of tiled images FCS Express will use information stored in the filename to piece the images back together. In particular the words "row" and "col" followed by a number indicate the position of the tile in the image. Below are a few examples of this naming convention:
mytiledimage_row00_col00.tif
mytiledimage_row00_col01.tif
mytiledimage_row00_col02.tif
The above file names represent the first three tiles (from left to right) of a tiled image as indicated by their "row" and "col" positions. The words "row" and "col" and the associated tile position should be added at the time of acquisition if possible or post acquisition by manually renaming the files or using a file renaming program.
Starting with a Large Image
Large images may also be analyzed by Cell Profiler and FCS Express. Due to the memory limits of most computers these images must first be "split" into smaller images for image segmentation. We recommend splitting larger images using Total Image Slicer to 500 x 500 pixel squares and setting the File name template as: [name]_row%row_col%col.tiff . (Total image slicer will simultaneously and automatically split the image to discrete squares and rename them with the "rowXX" "colXX" naming convention).
Exporting Tiled Images in CellProfiler for use in FCS Express
The CellProfiler modules that will be discussed in this section are Load Images, Convert Objects to Images, Save Images, and Export to Spreadsheet. The completed pipeline for this example can be found in the Tutorial Sample Data archive within the Exporting Tiled Images in CP folder is named CellProfilerTiledExportCOMPLETED.cpproj. The completed pipeline is meant to be used as a template for comparison to your pipeline and represents the finished product of this tutorial.
The example data set we will be using can be found in the Tutorial Sample Data archive within the Exporting Tiled Images in CP folder is named "In_Situ_Cerebellum_Staining". The sample data set (kindly provided by Dragan Maric, Ph.D) represents a 3 color + bright field experiment in which a portion of a rat cerebellum was imaged. To access the final product of the pipeline and a layout of the data please visit our Applications Examples webpage.
The steps for exporting data from CellProfiler have been broken down by module according to the CellProfilerTiledExport.cp pipeline. After loading the CellProfilerTiledExport.cpproj pipeline in CellProfiler, follow the steps below to amend the pipeline to prepare for export to FCS Express.
Selecting Default Output Folders
To organize your data correctly for FCS Express, ensure the Default Output Folder is the same folder where your images are stored and used for the Default Input Folder (Figure T24.42).
Note: The DefaultOUT.mat file will also be exported to the Output Folder. This file is for use in MATLAB. If you do not wish to use this file, we recommend deleting it to conserve disk space.

Figure T24.42 Set the Default Input and Output Folders to the Same Location
A. Loading Images, Extracting Metadata, Assigning Names
In order for FCS Express to recognize your data is in 96 well format, metadata from the image file names must be extracted. A Regular Expression must be defined to find the metadata in the file name or path of the data by following these steps in CellProfiler (Figure T24.2):
1.Click Images category in left column of CellProfiler window.
2.Drag and drop In_Situ_Rat_Cerebellum_Staining subfolder of Exporting Tiled Images in CP into designated area.
3.Click Metadata category directly below Images.
4.Click Yes radio button in the Extact metadata? box.
5.Confirm Metadata extraction drop-down list is set to Extract from file/folders.
6.Confirm Metadata source drop-down list is set to File name.
7.Click the magnifying glass icon to the right of Regular Expression to field.
8.Erase all text from Regex: field.
9. Copy and paste following text into Regex: field, which will define the row tile and column tile:
| _0000_350_DAPI_row(?P<rowtile>[0-9]{1,3})_col(?P<coltile>[0-9]{1,3}).tiff |
10. Click Submit in Regular expression editor window.
11. Click Update button in lower portion to see information extracted for each of the images.
12. Click NamesandTypes directly below Metadata.
13. Change Assign a name to drop-down list to Image matching rules.
14. Confirm CellMask in blank field to right of Contain drop-down list.
15. Confirm CellMask is entered in Name to assign these field.
16. Click Add another image button.
17. For second image set, enter GLAST in blank field to right of Contain drop-down list.
18. Confirm GLAST in Name to assign these field.
19. Click Add another image button.
20. For third image set, enter Bright in blank field to right of Contain drop-down list.
21. Confirm Bright in Name to assign these field.
22. Click Add another image button.
23. For fourth image set, enter DAPI in blank field to right of Contain drop-down list.
24. Confirm DAPI in Name to assign these field.
25. Click Update button in lower portion.
26. Screen should resemble Figure T24.43 below for Metadata module.
Notes:
•Follow these links for a tutorial on "Regular Expressions" and how to translate regular expressions.
•Setting up metadata and regular expressions only has to be done for one image in the set. For this example, it has been set up for the DAPI channel.
•The regular expression output names: Well, Row, Column, and Col may only be used with plate based or montage imaging experiments. Using any of these names for a single image experiment will result in a non-working export.
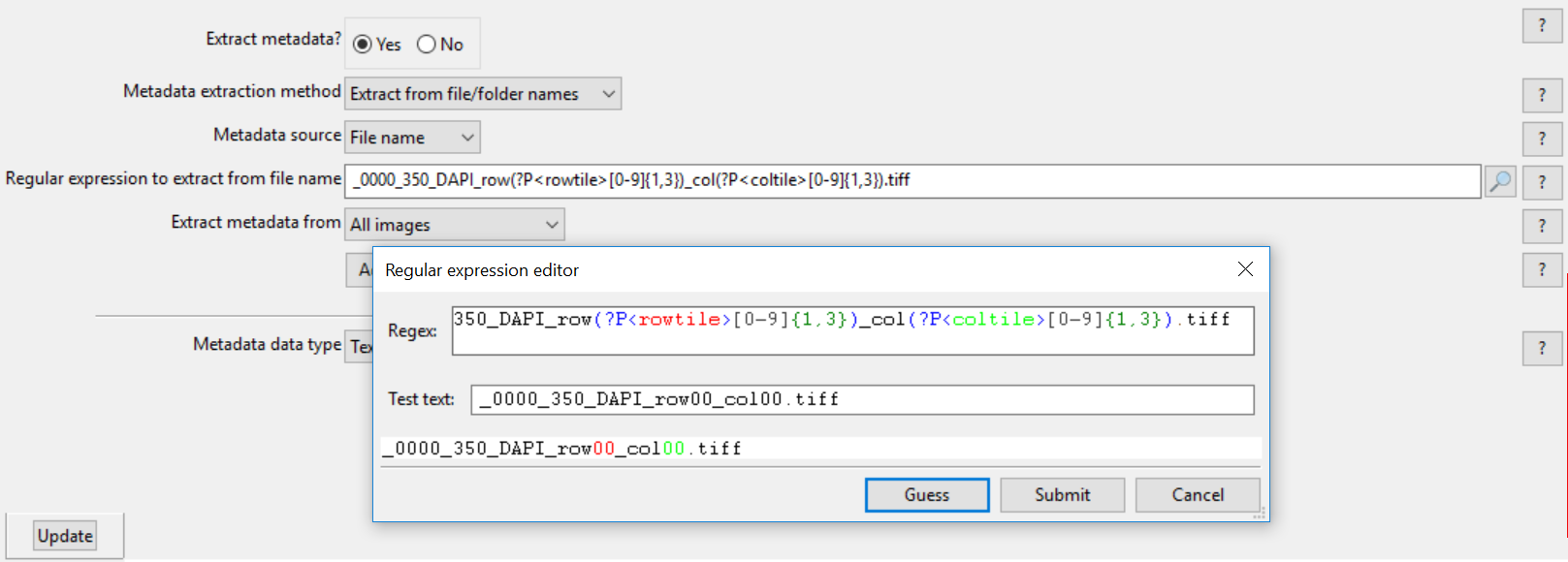
Figure T24.43 Setting up a Regular Expression in the LoadImages Module for a tiled image experiment.
B. ConvertObjectsToImage Modules
In these modules, the objects you have defined through the "Identify Primary/Secondary/Tertiary Objects" modules will be converted to image masks that FCS Express will use during import and analysis. See Figure T24.43 for an example of the module window.
1.Select the first ConvertObjectsToImage module in the pipeline.
2.Choose Nuclei from Select the input objects drop-down list.
3.Enter Nuclei in the Name the output image field.
4.Select uint 16 from the Select the color type drop-down list (see Figure T24.43 below).
5.Select the second ConvertObjectsToImage module in the pipeline.
6.Choose Cells from Select the input objects drop-down list.
7.Enter Cells in the Name the output image field.
8.Select uint 16 from the Select the color type drop-down list.
9. Select the third ConvertObjectsToImage module in the pipeline.
10. Choose Cytoplasm from Select the input objects drop-down list.
11. Enter Cytoplasm in the Name the output image field.
12. Select uint 16 from the Select the color type drop-down list.
Note: Repeat steps 1-4 for as many objects you have defined through the Identify Primary/Secondary Objects module when defining your own pipeline.
Note: In order for FCS Express to properly import the data the Name the output image name must be the same as the Select the input objects name and is case sensitive.
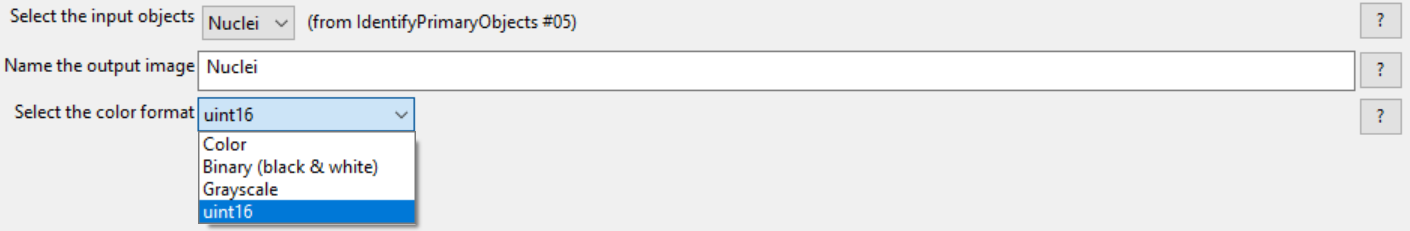
Figure T24.43 ConvertObjectsToImage Module: Defining Nuclei and Cells Image Masks
C. SaveImages Module
The image masks that were defined in the ConvertObjectsToImage modules must now be saved and related to the original images/data.
1.Select the first SaveImages module in the pipeline.
2.Set the Select the type of image to save drop-down list to Image.
3.Choose Cells for the Select the image to save drop-down list.
a.Note: when defining your own pipeline, select the appropriate image you defined in the ConvertObjectsToImage module.
4.Set the Select method for constructing file names drop-down list to Single name.
5.Enter CellLabel_ in the Enter single file name field.
6.Right-click after CellLabel_, choose rowtile, and enter an underscore "_"after it.
7.Right-click after rowtile_ and choose coltile.
8.Select the second SaveImages module in the pipeline.
9.Set the Select the type of image to save drop-down list to Image.
10. Choose Nuclei for the Select the image to save drop-down list.
11. Set the Select method for constructing file names drop-down list to Single name.
12. Enter NucleiLabel_ in the Enter single file name field.
13. Right-click after NucleiLabel_, choose rowtile, and enter an underscore "_"after it.
14. Right-click after rowtile_ and choose coltile.
15. Select the third SaveImages module in the pipeline.
16.Set the Select the type of image to save drop-down list to Image.
17. Choose Cytoplasm for the Select the image to save drop-down list.
18. Set the Select method for constructing file names drop-down list to Single name.
19. Enter CytoplasmLabel_ in the Enter single file name field.
20. Right-click after CytoplasmLabel_, choose rowtile, and enter an underscore "_"after it.
21. Right-click after rowtile_ and choose coltile.
Perform steps 22-27 on both SaveImages modules set up for CellLabel, NucleiLabel, and CytoplasmLabel .
22. Confirm tiff from the Saved file format drop-down list.
23. Set Image bit depth drop-down list to 16-bit integer.
24. Choose Default Output Folder from Output file location drop-down list.
25. Click Yes radio button in the Overwrite existing files without warning? box.
26. Set When to save as Every cycle from the drop-down list.
27. Click Yes radio button in Record the file and path information to the saved image? box.
Note: rowtile and coltile are values you defined as regular expressions for the load images module. Using regular expressions allows every output file to be assigned a unique file name based on the text from the input file name. This will prevent output file names from overwriting each other and allows FCS Express to identify the unique image mask associated with each image file.
The SaveImages module for CellLabel should now look like Figure T24.45.
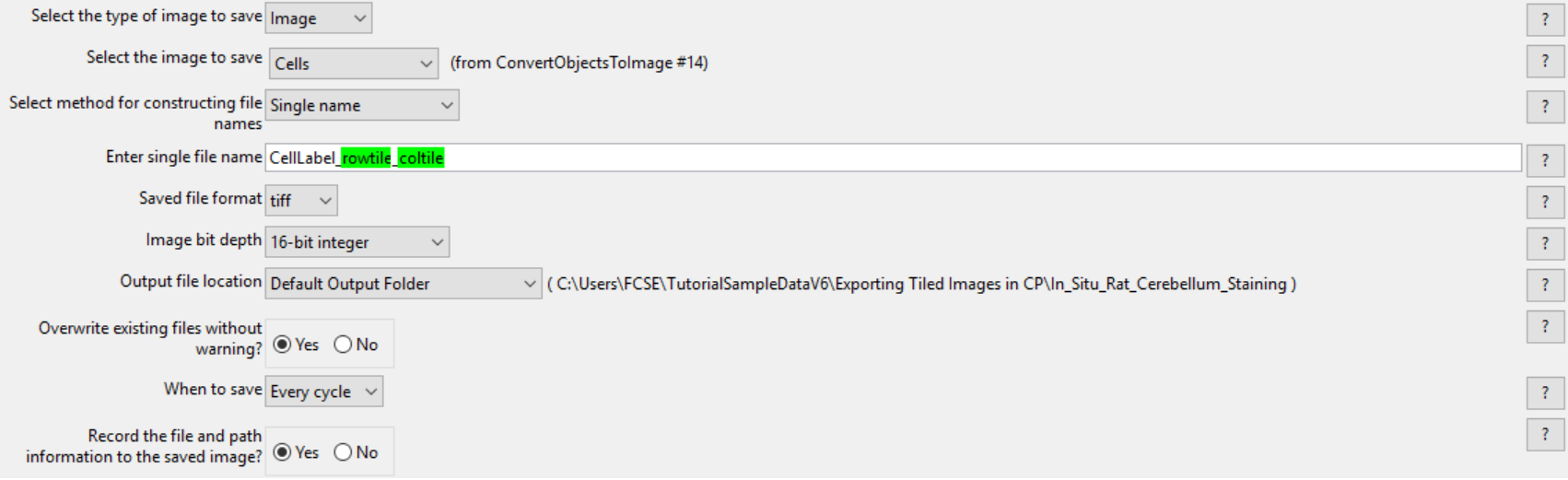
Figure T24.45 Defining the SaveImages Module
D. ExportToSpreadsheet Module
Now that all of the images and object image masks have been defined and saved for analysis, you must now ask CellProfiler to export the measurements you wish to view in FCS Express. For this example, we will export all measurements.
(Note: If you would like to only export certain parameters please see the Selecting Individual Parameters for CellProlifer Export section)
1.Select the first ExportToSpreadsheet module.
2.Confirm the Select the column delimiter drop-down list is set to Comma (",").
3.Choose Default Output Folder from the Output file location drop-down list.
4.Click No radio button in the Add a prefix to file names? box.
5.Click No radio button in the Export all measurement types? box.
6.Choose Image from the first Data to export drop-down list.
7.Click No radio button in the Use the object name for the file name? box.
8.Replace DATA.csv with Image.cptoc in File name field.
9.Select the second ExportToSpreadsheet module.
10.Confirm the Select the column delimiter drop-down list is set to Comma (",").
11.Choose Default Output Folder from the Output file location drop-down list.
12.Click No radio button in the Add a prefix to file names? box.
13.Click No radio button in the Export all measurement types? box.
Note: In this case, measurements have already been chosen for you and the nuclei.cpout file has been setup. Click on the Press to select measurements button to review which measurements have been selected.
14. Click Add another data set button.
15. Choose Cells from the second Data to export drop-down list.
16. Click No radio button in the Use the object name for the file name? box.
17. Replace DATA.csv with cells.cptoc in File name field.
18. Click Add another data set button.
19. Repeat steps 14-18 for "Cytoplasm" and set the file name as cytoplasm.cpout.
The second ExportToSpreadsheet module options should look like Figure T24.46 when you are done.
Note 1: on file extensions: ".cptoc" stands for CellProfiler Table of Contents and ".cpout" stands for CellProfiler Output. The "Image.cptoc" file stores information about the location and number of images for all objects processed in the pipeline, while the separate "Nuclei.cpout" and "Cells.cpout" store the actual raw data associated with individual objects and analysis. There should be a separate ".cpout" file defined for each object mask defined in CellProfiler. Only one "Image.cptoc" file is needed.
Note 2:The name of your .cpout file must begin with the name of the object data you are exporting and may only be include the object name and regular expressions. For example: the file names nuclei.cpout and nuclei_<regular express>.cpout will result in a proper export while dataset1nuclei.cpout or <regularexpression>_nuclei.cpout will resulting in a non-working export.

Figure T24.46 The second ExportToSpreadsheet Module Set Up to Export nuclei.cpout, cells.cpout, and cytoplasm.cpout.
20. Select Analyze images to run the pipeline. The folder where your images are stored will now contain the cells.cpout, nuclei.cpout, cytoplasm.cpout and Image.cptoc files. There will also be a CellLabel, NucleiLabel, and CytoplasmLabel image associated with every original image/well from the tiled data set.
In the next section, we will import and analyze the Tiled or Montage Image.
