Analyzing and Exporting Multi Channel Image Results with an .ijx file
In this section you will learn how to analyze and export a group of multi channel image results from ImageJ using the DNSMacroExample and an .ijx analysis configuration file, which was created in the previous section with the macro-enabled Excel worksheet* or manually edited in ImageJ or Excel.
We will be using the macros and images from the tutorial sample data set→ImageJ Tutorial folder. The DNSMacroExample.ijm macro will be used along with the images found in the Multi Channel Analysis folder (Images 1 and 2 for the DAPI and FITC channels).
| 1. | Open ImageJ. |
| 2. | Install the DNSMacroExample in ImageJ (Note: if the DNSMacroExample has already been installed you may skip step 2). |
When this macro file is installed in ImageJ (Plugins→Macros→Install), a red rectangle with three color blobs button with the hint "De Novo Software Exporter" will be shown in the toolbar (figure T28.113).
Figure T28.113. The ImageJ tool bar with the DNSMacroExample.ijm macro installed. Notice the red rectangle with three color blobs button that indicates the De Novo Software Exporter.
| 3. | Click on the De Novo Software Exporter button (red rectangle with three color blobs) in the Image J tool bar (figure T28.113). |
| 4. | Choose From an Analysis Configuration File (.ijx) (figure T28.114). |
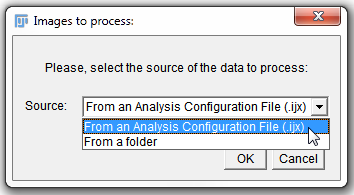
Figure T28.114 Choosing an .ijx file to process images.
| 5. | Click OK. |
| 6. | Choose either DeNovo.ijx (if your .ijx file was created with the macro-enabled Excel worksheet) or FileListForDnsExporter.ijx (if your .ijx file was manually edited in ImageJ or Excel) from the Multi Channel Image Experiment folder within the Tutorial Sample Data folder→ImageJ Tutorial directory folder. |
| 7. | Click Open. |
| 8. | Choose dapi as the Channel. |
Choosing the channel in step 8 will define which channel should be used for image segmentation. In this case we are using the dapi stain which is defining the nuclear area of the cells for segmentation.
| 9. | Click OK. |
ImageJ will now process all of the files listed in the .ijx Analysis Configuration file and create a .ijout file for each set of two channel images and a folder for each of the matching .results.txt and .mask.tif files.
We will now open the exported data with FCS Express.
| 10. | Open FCS Express 4 Image Cytometry. |
| 11. | Click on the Insert Tab→2D Plots→Dot. |
| 12. | Click anywhere on the layout to insert the Dot plot. |
| 13. | Choose ImageJ Files (*ijout) from the Files of type drop down menu. |
| 14. | Navigate to the Tutorial Sample Data folder→ImageJ Tutorial→Multi Channel Image Experiment→Multi Channel Analysis folder. |
| 15. | Choose Image1DAPI1.ijout. |
| 16. | Click Open. |
| 17. | Click on the Insert Tab→2D Plots→Picture Plot. |
| 18. | Click anywhere on the layout to insert the picture plot. |
The layout should now contain a Dot plot and Picture Plot for the data exported from ImageJ (figure T28.115). Using the Next and Previous buttons from the Data Tab→Change Data on All Objects section will change between the data from the first and second images that were processed. By clicking on the Channel Label in the picture plot (DAPI) you will be able to choose between the DAPI and FITC channels (Figure T28.115 right). Please see the further resources section below for more information about working with plots and imaging data in FCS Express.
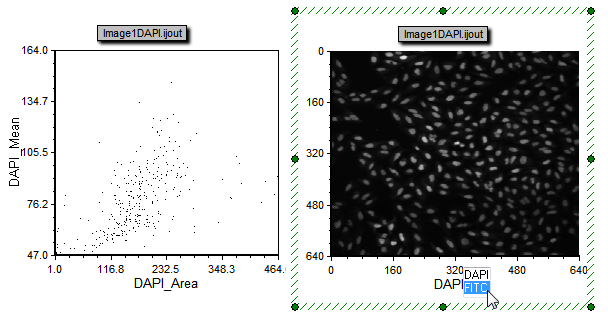
Figure T28.115 A dot plot and picture plot created from the .ijout files that were exported using the .ijx analysis configuration file. Since there are multiple channels you can change the axis of the picture plot to choose which channel to view.
Further Resources:
*macro written by Peter Haub
