It is often useful to look at many samples on the same plot to see sample variation and to compare experimental samples against a control. In this section, you will learn how to overlay multiple kinetics samples, apply parameter math to the overlays, and format the overlays to make the data easier to view.
Although any kinetics data files of your own can be used to follow along, you have the option of using the same sample data files used in this tutorial. The sample data files are located in the Tutorial Sample Data archive. The KineticsTutorialFormatting.fey, kinetics8.008, kinetics9.009, kinetics10.010, and kinetics25.025 data files will be used here.
| 1. | Select File tab→Open Layout. |
| 2. | Select the KineticsTutorialFormatting.fey layout file located in the FCS Express Sample Data directory from the Load Layout dialog. |
| 3. | Ctrl+Click on samples kinetics8.008, kinetics9.009, and kinetics10.010 to select them in the data list. |
| 4. | Drag the files from the data list onto the Kinetics Overlay plot. |
| 5. | Release the mouse button to drop them on the plot |
| 6. | Choose add the files to the plot as new overlays. |
| 8. | Change the Y-Axis parameter on the Kinetics Overlay plot to Ca++ Ratio. |
| 9. | Select the Live Cells gate from Gating→Create Gates→Current Gate. |
The plot now contains overlays for each sample and should look like Figure T21.19.
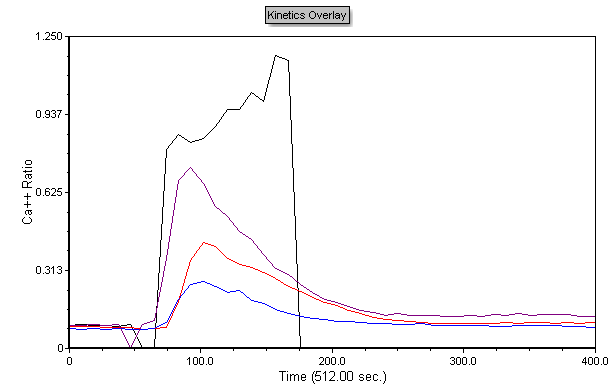
Figure T21.19 Multiple Kinetics Overlays
We will now format the plot to make it more visually appealing.
| 18. | Right-click on the plot. |
| 19. | Choose Format from the pop-up menu. |
| 20. | Choose the Overlays category. |
| 21. | Ctrl+Click on all of the plots in the list box to select them. |
| 22. | Change Line Options→Width to 1.50. |
| 23. | Choose the Legend category. |
| 24. | Click on the Visible check box to select it. |
The plot should now look like the Figure T21.20.
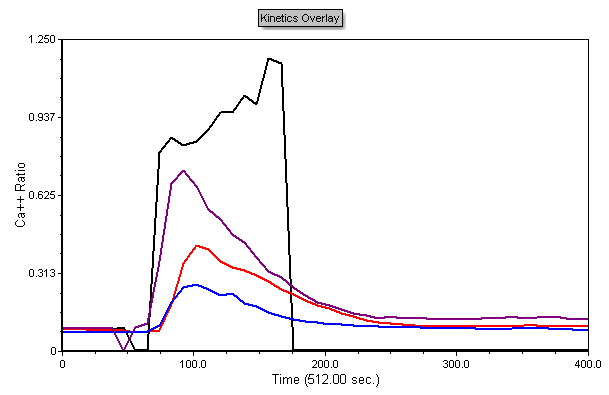
Figure T21.20 Kinetics Plots Formatted
Finally, we will change the resolution of a kinetics plot.


