The MultiCycle Tab
MultiCycle can automatically fit and analyze cell cycle histograms with multiple models. You can control the number and type of populations to fit, the currently displayed model, and the background options using the MultiCycle Tab (Figure 5.9). The Multicycle tab will only appear when a Multicycle DNA Analysis plot has been selected in the layout.
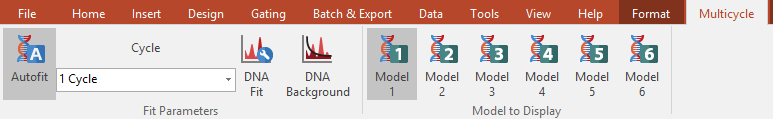
Figure 5.9 The Multicycle Tab
To analyze a DNA histogram automatically:
1. Insert a DNA Histogram onto your layout using the Insert→1D Plots→MultiCycle DNA command.
2. Select the DNA Histogram and verify that the MultiCycle→Fit Parameters→Autofit command is highlighted. With Autofit, MultiCycle will choose the Cycle parameter for you.
3. Right-click on the DNA Histogram and select Statistics→DNA Cycle Statistics from the pop-up menu. Adjust the position of the statistics window.
4. Right-click on the DNA Histogram and select Statistics→DNA Model Summary Statistics from the pop-up menu. Adjust the position of the statistics window.
The current model being displayed in the DNA Cycle Statistics is highlighted in the MultiCycle→Model to Display group. To change the model, click on one of the other five Model commands within the Model to Display group.
To change to a manual fit, click on MultiCycle→Fit Parameters→Autofit and verify that the command/button is not highlighted. With a manual fit:
•To change the number of populations for the fit, or the parameters for a 1 Cycle fit, use the MultiCycle→Fit Parameters→Cycle drop-down list.
•To change the Fit Model Properties, use the MultiCycle→Fit Parameters→DNA Fit command to bring up the DNA Fit options.
•To change background parameters, select the MultiCycle→Fit Parameters→DNA Background command to bring up the DNA Background options.
