Dimensionality Reduction
The table below describes the Dimensionality Reduction pipeline steps that are available in FCS Express. If you would like to recommend additional Dimensionality Reduction methods to be provided with FCS Express, please contact support@denovosoftware.com.
|
|
|---|---|
Step
|
Description
|
|
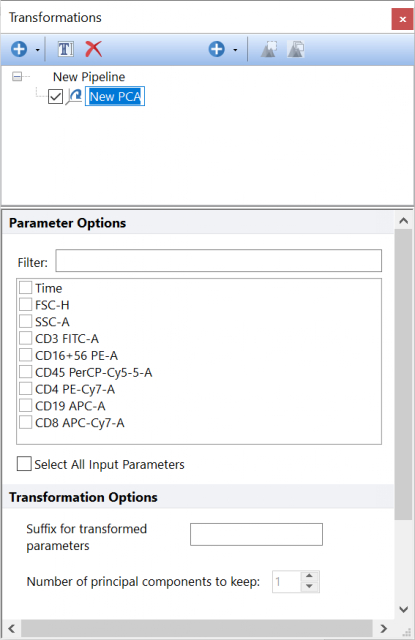
Calculates Principal Components of the input population, using the parameters selected in the Parameter Options list. Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The Number of Principal Components to display can be set in the Number of Principal Components to keep field.
The Suffix for transformed parameters filed allows to specify a suffix to add to all the parameters generated by this pipeline step. This is useful when multiple steps of the same type (e.g. multiple PCA steps) are present into the same pipeline.
|
|
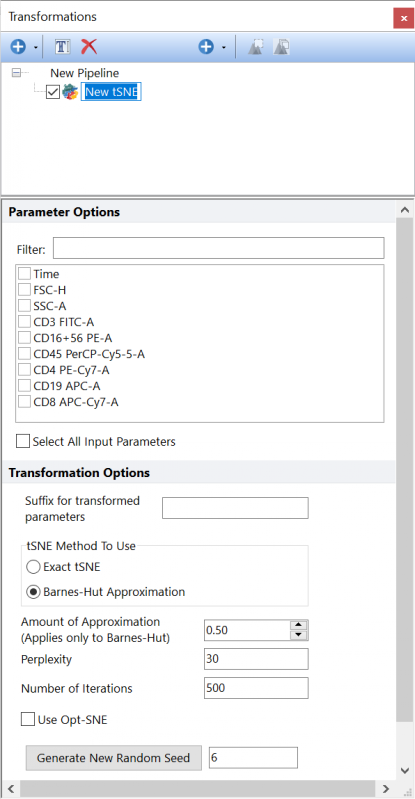
Performs dimensionality reduction using the tSNE algorithm on the input population, using the parameters selected in the Parameter Options list (see image below). Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The Suffix for transformed parameters filed allows to specify a suffix to add to all the parameters generated by this pipeline step. This is useful when multiple steps of the same type (e.g. multiple tSNE steps) are present into the same pipeline.
For additional detail on the tSNE algorithm please refer to the tSNE (viSNE) chapter of the manual.
The following settings may be customized:
•tSNE Method to Use •Amount of Approximation (Applies Only to Barnes-Hut) •Perplexity •Number of Iterations •Use Opt-SNE •Generate New Random Seed
Please refer to the Defining a tSNE transformation chapter for additional details on tSNE settings.
|
|
Performs dimensionality reduction using the UMAP (Uniform Manifold Approximation and Projection) algorithm on the input population, using the parameters selected in the Parameter Options list (see image below). Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The Suffix for transformed parameters filed allows to specify a suffix to add to all the parameters generated by this pipeline step. This is useful when multiple steps of the same type (e.g. multiple UMAP steps) are present into the same pipeline.
UMAP is a dimensionality reduction technique that allows users to create new UMAP X and UMAP Y parameters from a high-dimensional dataset. The two main steps of UMAP are: 1.Creation of an high-dimensional graph. A weighted graph in which a point is linked to its nearest neighbors. The amplitude of the neighborhood is a crucial parameter and is dictated by the Number of neighbors parameter (see below). 2.Creation of a low-dimensional (2 dimensional) graph as similar as possible to the high-dimensional graph resulting in UMAP X vs UMAP Y parameters.
The following settings may be customized by the user:
•Number of neighbors. Sets the number of approximate nearest neighbors used to create the initial high-dimension graph. Number of neighbors is a crucial parameter as low values will instruct the UMAP algorithm to focus more on local structure by constraining the number of neighboring points considered when analyzing the data in high dimensions, while high values will instruct the UMAP algorithm towards representing the overall structure while sacrificing fine detail. Minimum input is 2 and maximum input is 50. •Min Low Dim Distance. Sets the Minimum Distance between points in the low-dimensional map (i.e. in the UMAP map). By setting low values, points will be clustered closer together. Minimum input is 0.00001 and maximum input is 2. •Number of Iterations. The number of cycles that the UMAP algorithm performs to refine the results. This is sometimes called "Number of Epochs". Minimum number of iterations is 1 and Maximum number of iterations is 10,000. •Generate New Random Seed. The UMAP algorithm is stochastic. To make it reproducible, a fixed Seed may be set. If the same dataset and the setting are used, by retaining the same Random Seed value, the same result will be achieved. UMAP may be run multiple times with different Random Seeds to evaluate the stability and the consistency of the population separations.
For more details on UMAP algorithm, please refer to the following resources:
•Leland McInnes et al. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2018 arXiv:1802.03426 •Becht E. et al. Dimensionality reduction for visualizing single-cell data using UMAP Nat Biotechnol. 2018;10.1038 •Andy Coenen, Adam Pearce. Understanding UMAP. https://pair-code.github.io/understanding-umap/
|