Formatting a Proliferation Histogram
In the course of this section, we will :
•Learn how to adjust the proliferation overlays to better visualize the populations of interest.
•Learn how to produce the curve fitting statistics.
•Learn how to smooth the proliferation histogram.
•Learn how to use a batch process for analyzing multiple proliferation samples.
1.Open the "Proliferation Tutorial Formatting Section.fey" layout in the Proliferation Tutorial folder within the Tutorial Sample Data folder.
2.Click on the "Proliferation Data" page tab.
3.Double-click the proliferation plot. The Formatting dialog will appear, docked to the right (Figure T22.11).
4.Select 3 - Noise from the Overlay List (Figure T22.11, blue highlighted text).
5.Uncheck the box next to Visible (Figure T22.11, red outline).
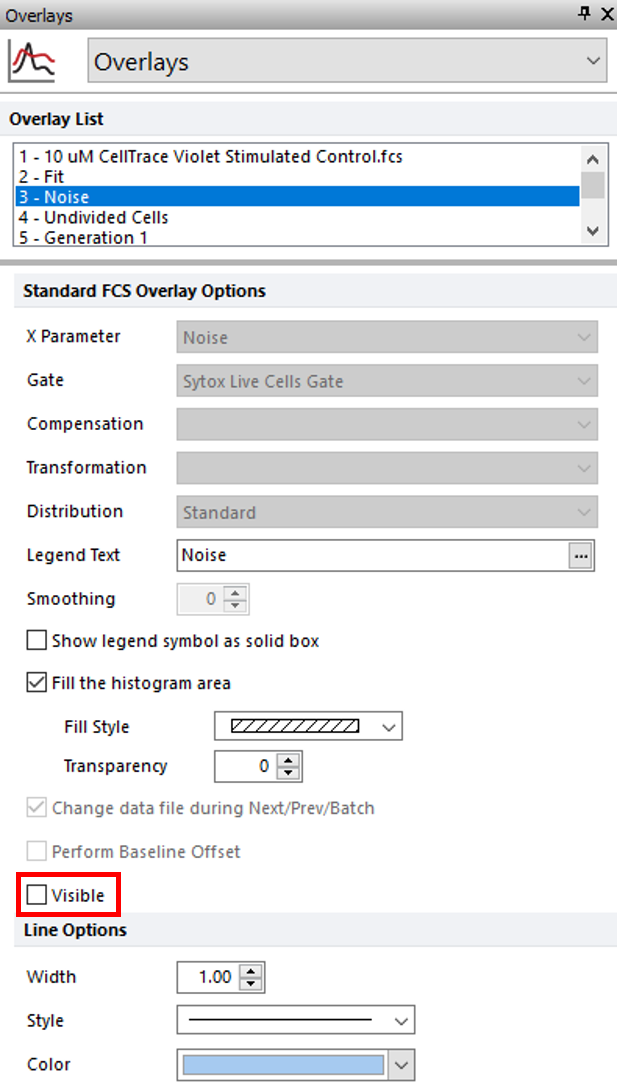
Figure T22.11 Removing Noise Overlay from Proliferation Plot
6.Select 10 - Generation 6 from the Overlay List (Figure T22.12, ![]() ).
).
7.For the Fill Style drop-down, select the first option for a completely filled rectangle (Fig. T22.12, ![]() ).
).
8.Change the Color to Blue (Figure. T22.12, ![]() )
)
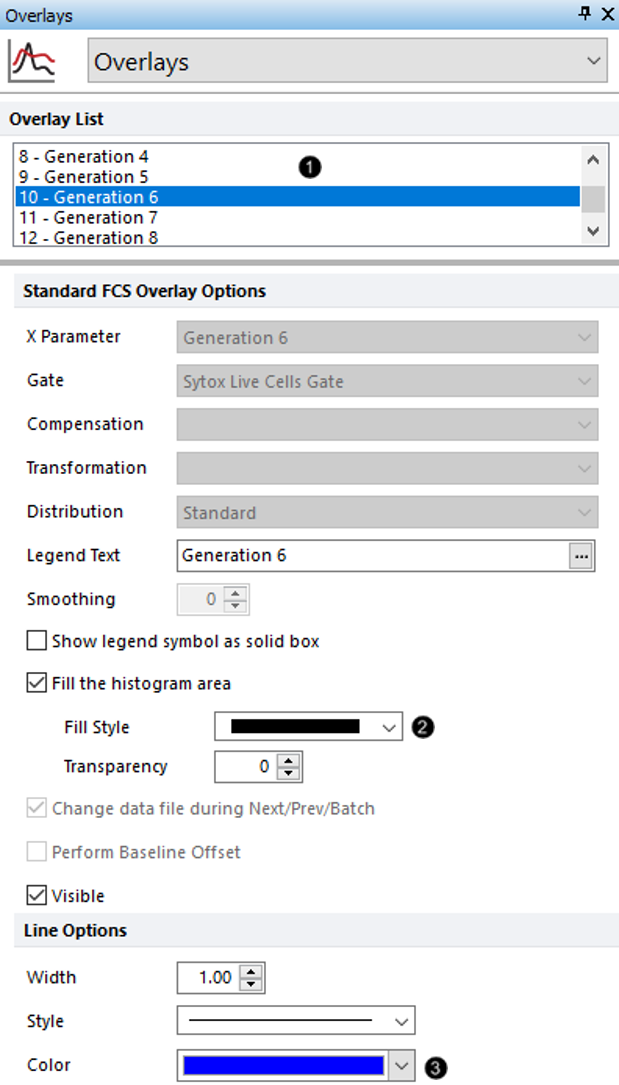
Figure T22.12 Changing Fill Style and Line Color for Generation 6 in Proliferation Plot
9.Select the 12 - Generation 8 from the Overlay List (Figure T22.13, ![]() ).
).
10. For the Fill Style drop-down, select the first option for a completely filled rectangle (Figure T22.13, ![]() ).
).
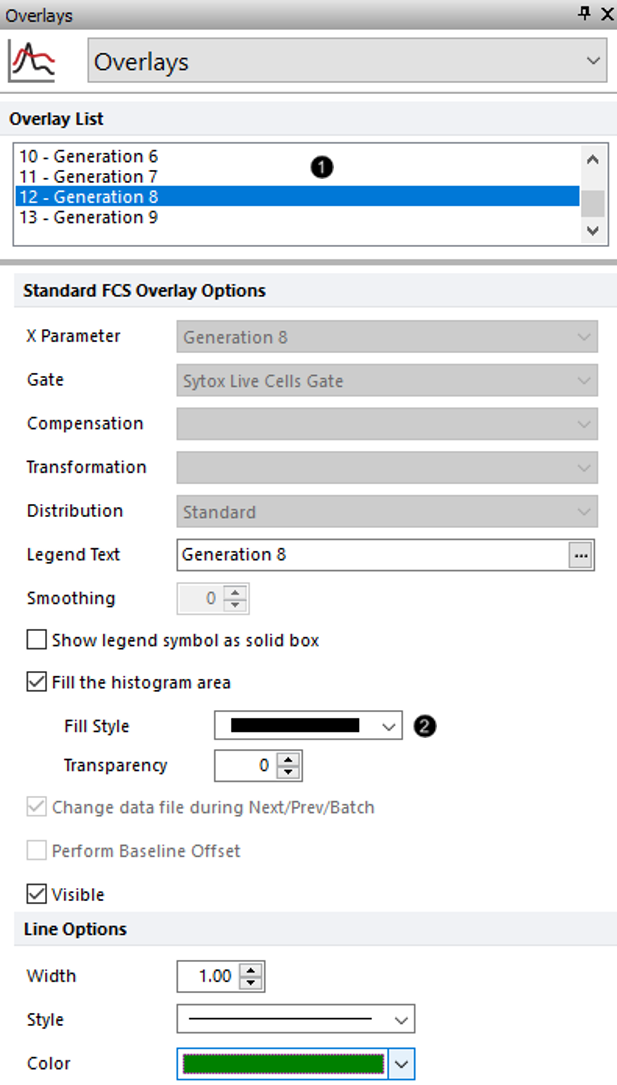
Figure T22.13 Changing Fill Style for Generation 8 in Proliferation Plot
The proliferation histogram now appears without displaying the noise, with the peak corresponding to generation 6 filled in blue, and the peak corresponding to generation 8 filled in green (Figure T22.14). This allows for easier confirmation and visualization of each generation and fitted peak. You can change the formatting of any and all of the different fitted elements and overlays in this way.
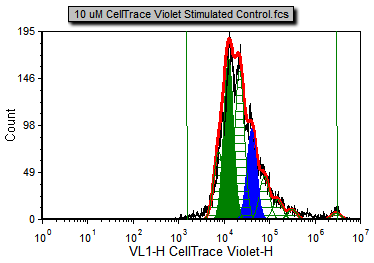
Figure T22.14 Proliferation Plot after Removing Noise and Changing Fill Style and Color for Two Generations in Overlays
Both the RMS error and R-Chi Square are measures of the goodness of the fit. When setting the proliferation model fit parameters, it is important to check how changing these parameters affect these two statistics. The combination of settings that provides the least RMS error and R-Chi Square is mathematically the best fit. We will now show the associated statistics for this proliferation model.
12. Right-click the proliferation plot.
13. Select Statistics→Proliferation Fit Statistics from the popup menu.
The Proliferation Fit Statistics window will now appear.
14. Right-click the proliferation plot.
15. Select Statistics→Proliferation Population Statistics.
The Proliferation Population Statistics window will now appear.
16. Move both statistic tables so that the proliferation histogram and tables are all visible as in Figure T22.15.
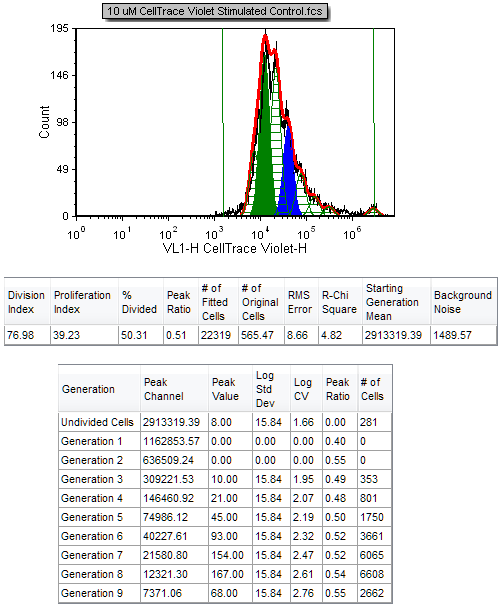
Figure T22.15 Proliferation Histogram with the Proliferation Fit Statistics and Proliferation Population Statistics
We now will smooth the proliferation histogram.
19. Click the proliferation plot to select it.
20. In Formatting dialog, select 1 - 10um CellTrace Violet Stimulated Control.fcs from the Overlay List.
21. Increase the Smoothing to 6.
The Proliferation histogram and associated statistics are updated immediately to reflect the smoothing (Figure T22.16). The black contour of the raw data is much smoother, but the fit changes significantly (note that the proliferation fit and proliferation population statistics have changed). Although this provides for a nicer visual presentation, this option should be used with caution since it affects the statistics obtained when using the raw data, and can thus affect the interpretation of the data. Note that the R-Chi Square value has increased from 4.82 to 17.76 as a result of smoothing the raw data, indicating an inferior fit. Thus it is advisable to cull statistics from an unsmoothed (raw) proliferation plot, while applying smoothing to a duplicate Proliferation plot or regular histogram for visual presentation only, if desired.
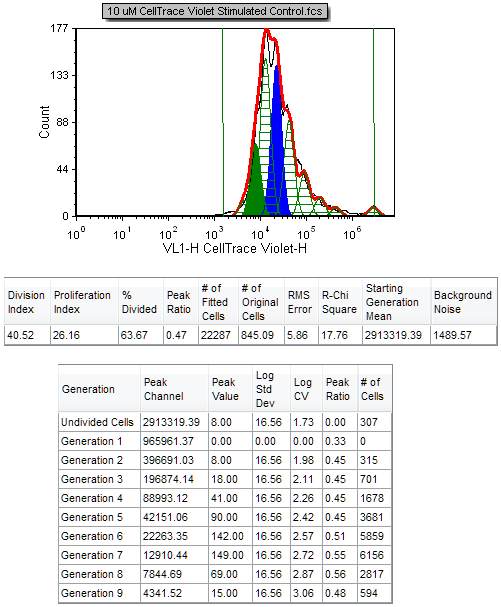
Figure T22.16 Proliferation Histogram and Associated Statistics with Smoothing
Now, we have formatted our plots for the experiment and inserted proliferation statistics, so we will create a batch process in the next section to summarize the three-sample experiment.
