Importing Single Image Data
This section pertains to data from a set of single images that were not obtained from a 96 well plate, such as images obtained from a slide.
After adjusting the export settings in CellProfiler and running your pipeline, you may proceed to analyze your images and import the data into FCS Express.
Importing the Single Image Experiment to FCS Express
Note on FCS Express Preferences before starting these steps of the tutorial:
The File selection dialog must be enabled:
A.Open the FCS Express User Options by choosing File→Options.
B.Check the When creating a new plot, always display the selection dialog box in the Files/Directories category.
(Enabling this feature is not a requirement of FCS Express, it has been selected for convenience in this tutorial.)
1. Load the Section2pipelineCOMPLETED.cpproj pipeline into CellProfiler 3.0.
2. Drag and drop the folder called 5 and 10 minute time point images folder into Images module.
3. Confirm the default input and output folders in CellProfiler to load and store the data as shown in Figure T26.14.
4. Click the Analyze Images button in the lower left of the in CellProfiler window.
5. Open FCS Express.
6. Open the FCS Express User Options by choosing File→Options
7. Select CellProfiler Options from the Data Loading category (Figure T26.19, orange highlighted text).
8. Select csv from the delimiter drop-down list.
9. Check the Load Images check box.
10. Uncheck the Load path names from Cell Profiler table of contents file check box.
11. Click OK to close the FCS Express User Options dialog.
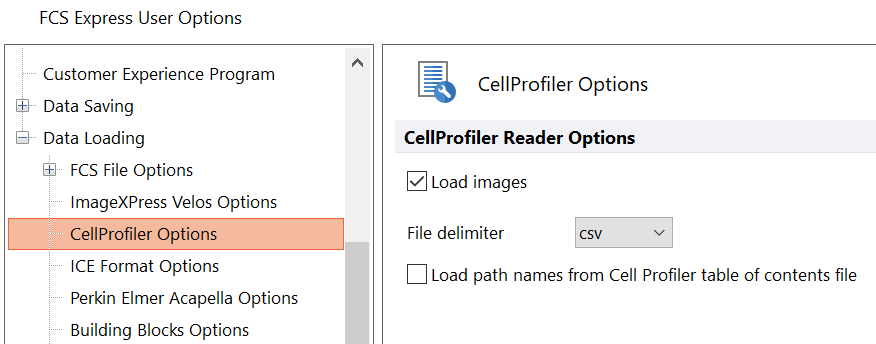
Figure T24.19 CellProfiler Data Loading Options
12. Select the Insert tab→Other Plots groups→Picture Plot command (Figure T26.20).
Figure T26.20 Inserting Picture Plot Insert Tab on Ribbon
13. Click anywhere on the layout and the Select data file dialog will appear.
14. Browse to the Default Output Folder where you stored your output data from CellProfiler.
15. Change the Files of type: drop-down list to CellProfiler (*.cpout) (Figure T26.21). Icons for the documents outputted by CellProfiler will appear. For this example, they will be labeled cells_05.cpout and nuclei_05.cpout.
16.Highlight cells_05.cpout and click Open.
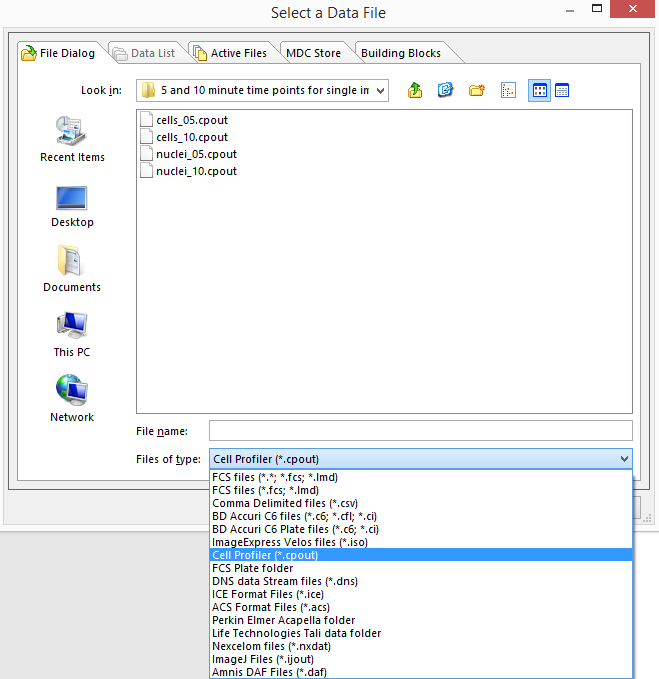
Figure T26.21 Select Data File Dialog
Note: For this example, we will use cells_05.cpout which will display data from the 5 minute time point objects classified in the IdentifySecondaryObjects module of CellProfiler which corresponds to the entire cell area. Nuclei_05.cpout will display data from the 5 minute time point objects classified in the IdentifyPrimaryObjects module of CellProfiler which corresponds only to the nuclear area of the cells.
17. Highlight Cytoplasm from the Image Parameter list box of the Open 2D Plots dialog (Figure T26.22) to view the cytoplasmic image.
18. Click OK.
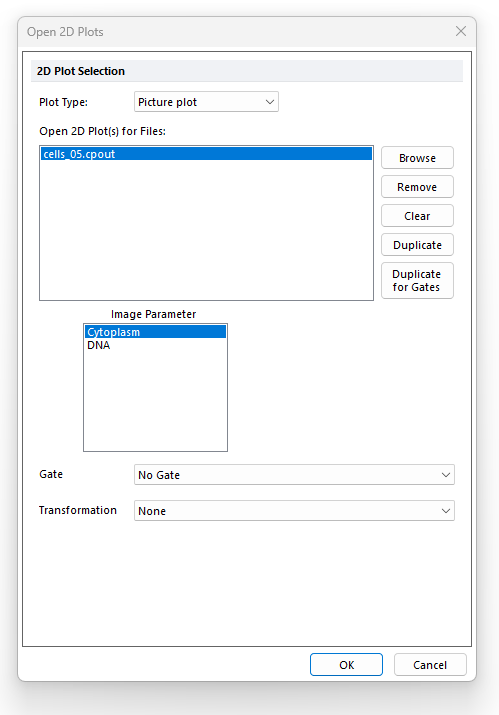
Figure T26.22 Choosing Image Parameters from the Open 2D Plots Dialog
A Picture Plot will appear on your layout, indicating that the data was successfully imported (Figure T26.23).
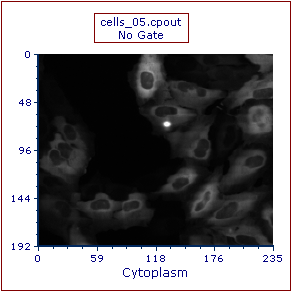
Figure T26.23 Example of the Cells (5 Minute Time Point) Picture Plot
Note about panning and zooming (please refer the working with picture plots chapter for more details)
By default, Picture plots display images at their original zoom starting from their top-right corner. When the original size of the picture exceeds the size of the plot, to see a larger part of the picture the user can:
•enlarge the plot itself and/or (Figure T26.23a below)
•change the zoom of the Picture plot by selecting Specific Options from the Format tab of the ribbon bar (Figure T26.23b below). Should the zooming not being enough, the plot can be resized in addition.
If a different region of the image should be displayed within the picture plot, please change the panning (please refer the working with picture plots chapter for more details).
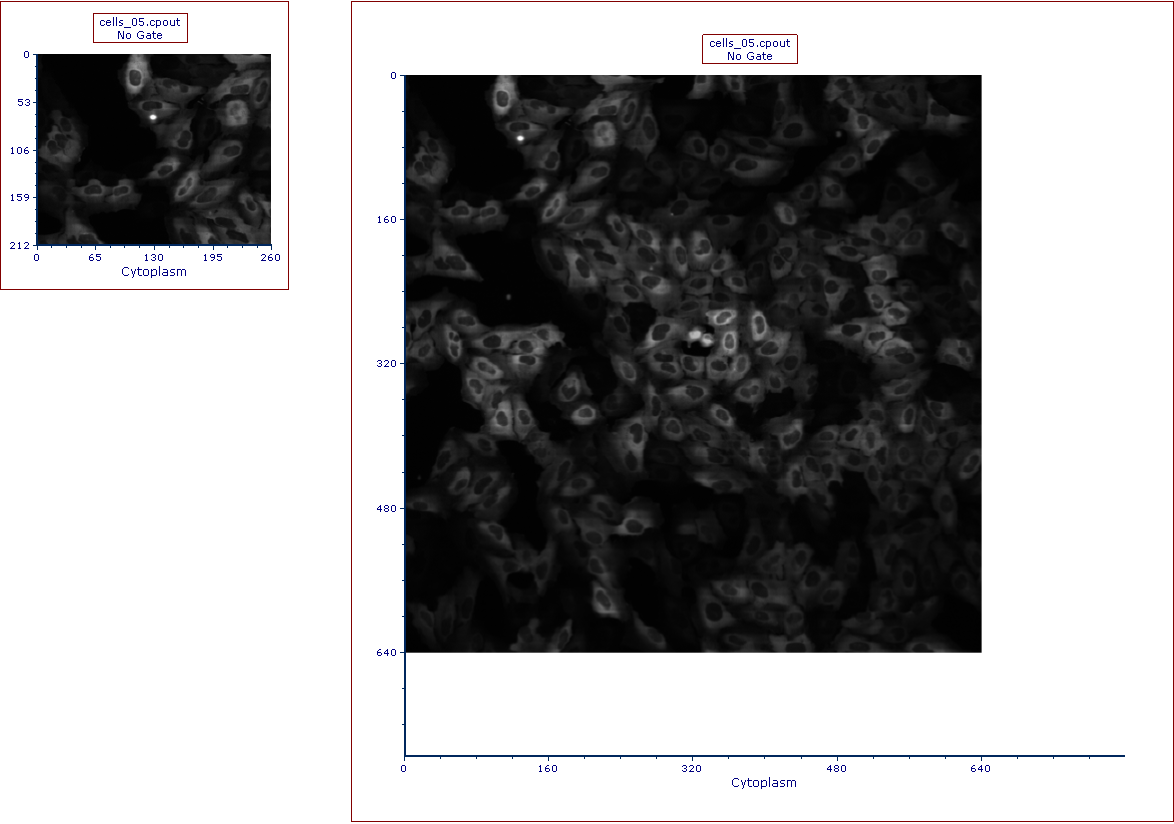
Figure T26.23a
The picture plot (left) has been resized in order to display the full picture (right)
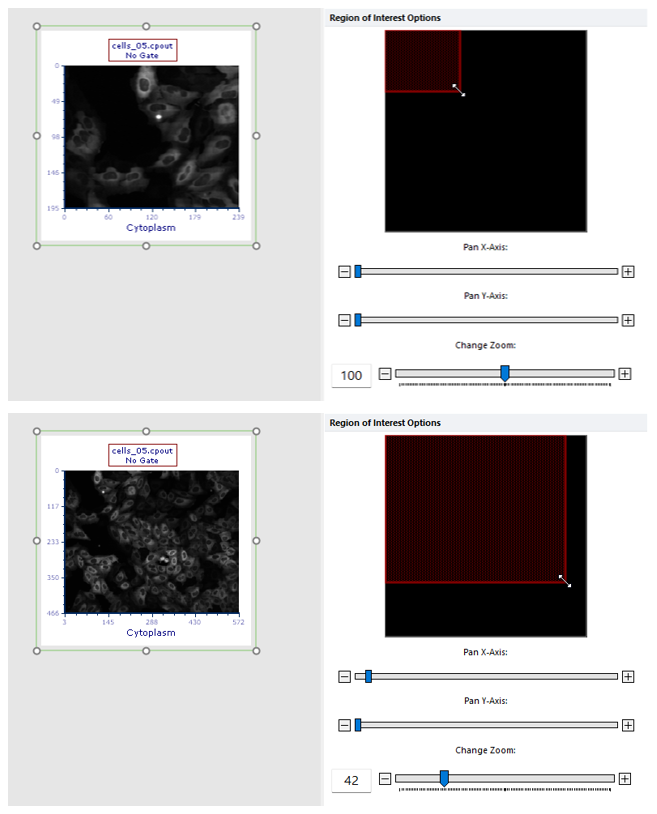
Figure T26.23b
The picture plot (top) has been zoomed through the Specific Options dialog in order to display a larger part of the picture (bottom).
19. Repeat steps 13-18 but highlight DNA as Image Parameter in the Open 2D Plots dialog.
Note: alternatively, you can duplicate the 2D plot previously created and then change the visualized parameter by clicking on the X axis and choosing DNA instead of Cytoplasm.
Zoom out on the plot or resize the plot area to display the entire picture in each plot, the layout should resemble Figure T26.24.
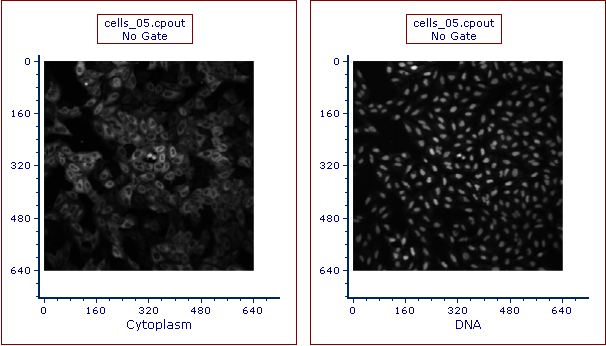
Figure T26.24 Picture Plots for cells_05.cpout for Cytoplasm and DNA Channels
Any type of 2D plot can be created by following the previous step. For training purpose, the user can try opening two Color Dot Plots of the same file (i.e. cell_05.cpout) depicting:
•AreaShape_Area with Intensity_MeanIntensity_Cytoplasm
•AreaShape_Area with Intensity_MeanIntensity_DNA
Tips: dot size on Dot plots and Color Dot plots, can be adjusted via the Overlay formatting dialog.
The layout should resemble Figure T26.25.
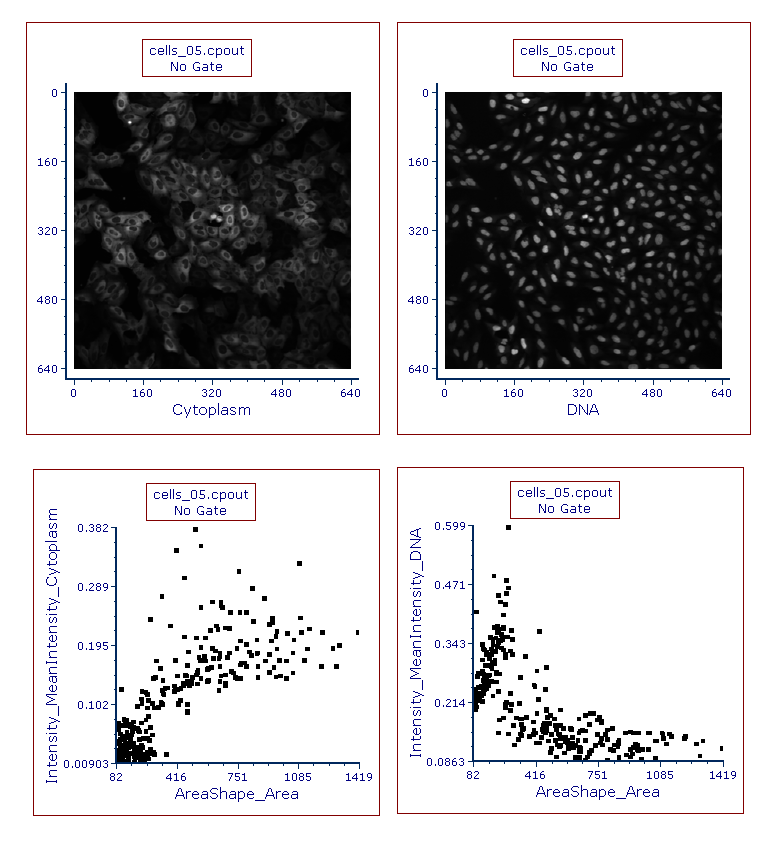
Figure T26.25 Picture and Color Dot Plots for cell_05.cpout
Any 2D gate can easily be created on the Picture Plots and on the Color Dot Plots as well. Inserting a new 2D plot gated on the picture plot gate will allow you to view the data as familiar dot plots, or view gated portions of the image (Figure T26.26).
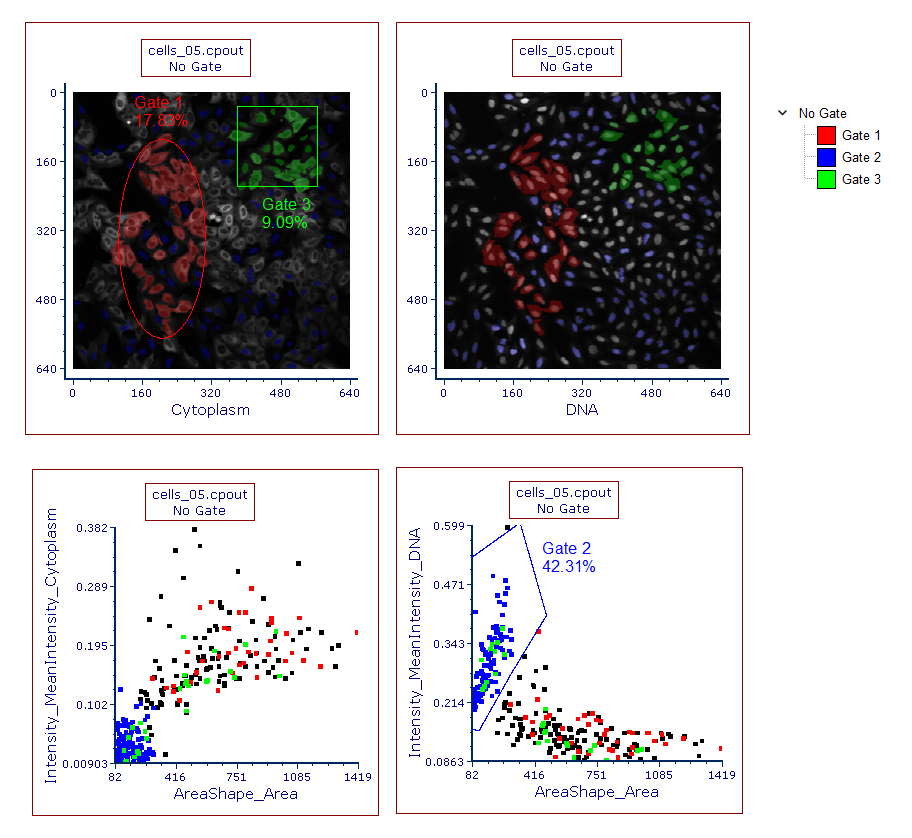
Figure T26.26 Example of Gating on a Picture Plot and Backgating from a Color Dot Plot to a Picture Plot
In the next exercise, we will export numeric data only from a 96 well plate.
