Quality Controls
The table below describes the Quality Controls pipeline steps that are available in FCS Express. If you would like to recommend additional Quality Controls methods to be provided with FCS Express, please contact support@denovosoftware.com.
|
|
|---|---|
Step
|
Description
|
|
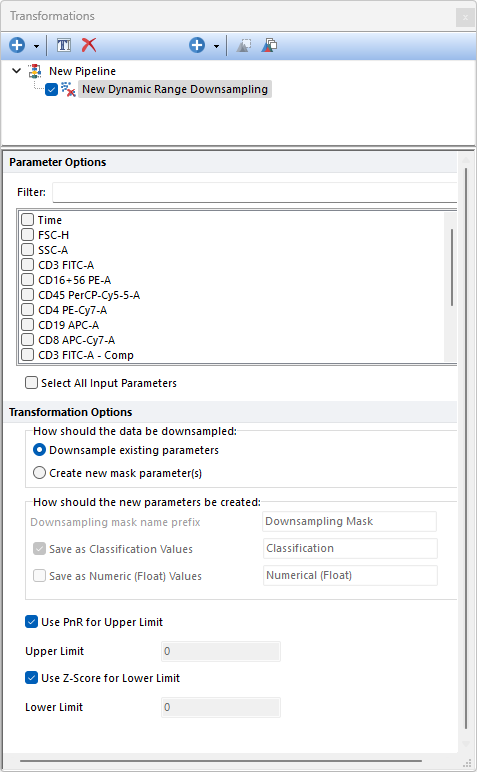
Dynamic Range Downsampling allows users to remove events that are equal to, or higher than, the defined upper limit, and/or to remove events that are equal to, or lower than, the defined lower limit.
Downsampling is performed parameter-by-parameter among the selected input parameters, on the population selected in the main pipeline body.
Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
Note: The Dynamic Range Downsampling method is used by the FlowAI pre-defined step to perform the "Dynamic Range check". When the step is run independently from the FlowAI algorithm, the Dynamic Range Downsampling step should be run on Linearly-scaled non-compensated data (please see the FlowAI pre-defined step section for further details on how to use Linearly-scaled non-compensated data as input data).
The following options are available: •Use PnR for Upper Limit: When checked, the range (i.e. the $PnR keyword) of the selected parameter(s) will be used as the upper limit. When unchecked, an Upper Limit field becomes active and allows a user defined upper range. •Use Z-Score for Lower Limit: When checked, the Lower Limit will be set to be the Z-Score method published in the original FlowAI paper (please see the FlowAI pre-defined step for more details and the bibliographic reference). When unchecked, a Lower Limit field becomes active and allows a user defined the lower range.
The How should the data be downsampled option allows two choices: oDownsampling Existing Parameters. No mask parameter is created and only the downsampled events will be available downstream this pipeline step. oCreate new mask parameters(s). One (or two; see below) new parameter is created. The name and the nature of this parameter can be customized with the How should the new parameter be created option (see below).
The How should the new parameters be created option allows to: oDefine the Downsampling mask names prefix. This is defaulted to Downsampling Mask but can be edited. Editing this prefix is especially useful to distinguish mask parameters generated by different downsampling pipeline steps. oDefine whether the new parameter should contain data saved as Classification Values. When this option is used, sampled events will be assigned with a "Include" value in that parameter while unsampled events will be assigned with a "Exclude" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter. oDefine whether the new parameter should contain data saved as Numeric (Float) Values. When this option is used, sampled events will be assigned with a "1" value in that parameter while unsampled events will be assigned with a "0" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter.
Note: when both the Save as Classification Values and the Save as Numeric (Float) value options are used, two mask parameters will be created.
|
|
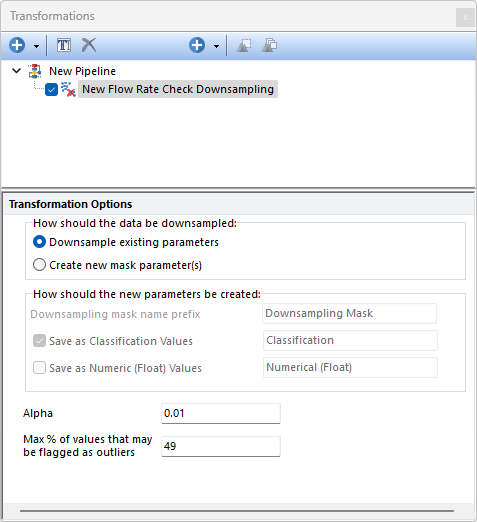
With the Flow Rate Check Downsampling step the steadiness of the flow (i.e. the number of events acquired per unit of time) can be checked. The flow rate is reconstructed using the Time parameter and the $TIMESTEP keyword from the data file (which is contained in all standard FCS file with version 3.0 or higher). If the $TIMESTEP keyword is not available, a default timestep of 1/10 second will be used to recreate the flow rate. An anomaly detection algorithm (built upon the generalized Extreme Studentized Deviate (ESD) test) detects and removes the data acquired during flow rate surges and shifts from the median value.
The algorithm is run parameter-by-parameter on the population selected in the main pipeline body.
The Flow Rate Check Downsampling algorithms integrated in FCS Express is based on the Flow Rate Check algorithm reported in the original FlowAI paper (G. Monaco et al, flowAI: automatic and interactive anomaly discerning tools for flow cytometry data, Bioinformatics, Volume 32, Issue 16, 2016) and is indeed used by the FlowAI pre-defined step to perform the Flow Rate Check.
The algorithms allows two inputs from the user: •Alpha: The the level of statistical significance used to accept anomalies detected by the ESD method. The default value is 0.01. The highest accepted value is 0.1. •Max % of values that may be flagged as outliers: The highest percentage of events allowed to be flagged as outliers. The highest accepted value is 49%.
The How should the data be downsampled option allows two choices: oDownsampling Existing Parameters. No mask parameter is created and only the downsampled events will be available downstream this pipeline step. oCreate new mask parameters(s). One (or two; see below) new parameter is created. The name and the nature of this parameter can be customized with the How should the new parameter be created option (see below).
The How should the new parameters be created option allows to: oDefine the Downsampling mask names prefix. This is defaulted to Downsampling Mask but can be edited. Editing this prefix is especially useful to distinguish mask parameters generated by different downsampling pipeline steps. oDefine whether the new parameter should contain data saved as Classification Values. When this option is used, sampled events will be assigned with a "Include" value in that parameter while unsampled events will be assigned with a "Exclude" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter. oDefine whether the new parameter should contain data saved as Numeric (Float) Values. When this option is used, sampled events will be assigned with a "1" value in that parameter while unsampled events will be assigned with a "0" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter.
Note: when both the Save as Classification Values and the Save as Numeric (Float) value options are used, two mask parameters will be created.
|
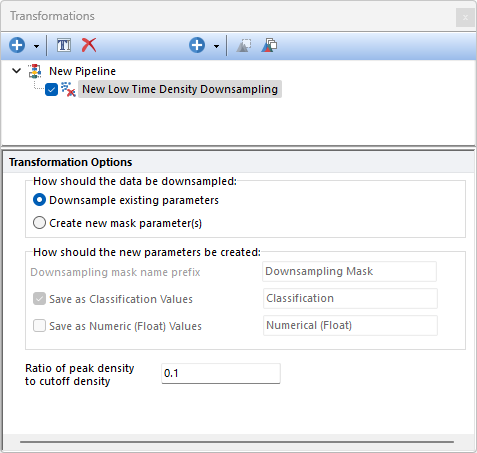
The Low Time Density Downsampling step checks the steadiness of the flow rate (i.e. the number of events acquired per unit of time). The Low Time Density Downsampling first reconstructs the flow rate using a kernel density estimation function that calculate the events density along the time. The highest density (i.e. the peak) is then calculated and events with a density lower than the threshold specified by the user are removed. The threshold is defined using the "Ratio of peak density value to cut off density value" field. The default value is 0.1, which means that events with a density lower than, or equal to, peak*0.1 (i.e. 10% of the peak) will be removed.
When the peak value is very high because of a spike in the Time channel, most of the events would be removed. To avoid this, when a spike in the Time channel is detected, the Low Time Density Downsampling does not remove any events.
The How should the data be downsampled option allows two choices: oDownsampling Existing Parameters. No mask parameter is created and only the downsampled events will be available downstream this pipeline step. oCreate new mask parameters(s). One (or two; see below) new parameter is created. The name and the nature of this parameter can be customized with the How should the new parameter be created option (see below).
The How should the new parameters be created option allows to: oDefine the Downsampling mask names prefix. This is defaulted to Downsampling Mask but can be edited. Editing this prefix is especially useful to distinguish mask parameters generated by different downsampling pipeline steps. oDefine whether the new parameter should contain data saved as Classification Values. When this option is used, sampled events will be assigned with a "Include" value in that parameter while unsampled events will be assigned with a "Exclude" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter. oDefine whether the new parameter should contain data saved as Numeric (Float) Values. When this option is used, sampled events will be assigned with a "1" value in that parameter while unsampled events will be assigned with a "0" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter.
Note: when both the Save as Classification Values and the Save as Numeric (Float) value options are used, two mask parameters will be created.
The Low Time Density Downsampling step integrated in FCS Express is based on the removeLowDensSections algorithm used in flowCut (J. Meskas et al, flowCut: An R package for precise and accurate automated removal of outlier events and flagging of files based on time versus fluorescence analysis, bioRxiv, doi: https://doi.org/10.1101/2020.04.23.058545) and is also used by the FlowCut pre-defined step.
|
|
|
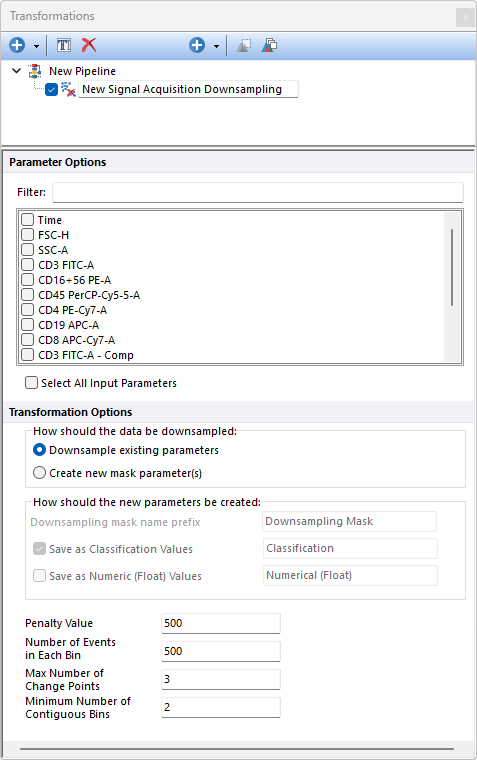
Provided that the signal intensity of a given population (i.e. median and variance of intensity) should be constant during acquisition in the vast majority of flow cytometry experiments (kinetics experiments are an exception to the rule), changes in the median and/or in the variance of the signal are usually indicators of issues with fluidics or other problems when acquiring data.
Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
Note: The Signal Acquisition Downsampling method is used by the FlowAI pre-defined step to perform the "Signal Acquisition check". When the step is run independently from the FlowAI algorithm, the Signal Acquisition Downsampling step should be run on Linearly-scaled on-compensated data (please see the FlowAI pre-defined step section for further details on how to use Linearly-scaled non-compensated data as input data).
When using Signal Acquisition Downsampling, the median and the variance of the signal of equally-sized bins of events acquired sequentially are calculated parameter-by-parameter on the population selected in the main pipeline body. The number of sequentially events in each bin is defined by the variables entered by the user (see below).
The Signal Acquisition Downsampling algorithm detects shifts in the median and/or in the variance of the fluorescence intensity between bins in each of the input parameters by using the Binary Segmentation algorithm of the changepoint R package.
The algorithm allows users to customize the following options: •Penalty Value: The value of the penalty for the changepoint detection algorithm. The higher the penalty value the less strict is the detection of the anomalies. The default is 500. •Number of Events in Each Bin: The value defines the size of each bin, i.e. the number of sequential events that each bin should contain. •Max Number of Change Points: The maximum number of changepoints (i.e. changes in the median and or in the variance intensify) that can be detected for each channel. The default is 3. •Min Number of Contiguous Bins: The minimum segment length (i.e. the number of bins between changes). The default is the minimum allowed by theory, i.e. 2.
The How should the data be downsampled option allows two choices: oDownsampling Existing Parameters. No mask parameter is created and only the downsampled events will be available downstream this pipeline step. oCreate new mask parameters(s). One (or two; see below) new parameter is created. The name and the nature of this parameter can be customized with the How should the new parameter be created option (see below).
The How should the new parameters be created option allows to: oDefine the Downsampling mask names prefix. This is defaulted to Downsampling Mask but can be edited. Editing this prefix is especially useful to distinguish mask parameters generated by different downsampling pipeline steps. oDefine whether the new parameter should contain data saved as Classification Values. When this option is used, sampled events will be assigned with a "Include" value in that parameter while unsampled events will be assigned with a "Exclude" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter. oDefine whether the new parameter should contain data saved as Numeric (Float) Values. When this option is used, sampled events will be assigned with a "1" value in that parameter while unsampled events will be assigned with a "0" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter.
Note: when both the Save as Classification Values and the Save as Numeric (Float) value options are used, two mask parameters will be created.
|
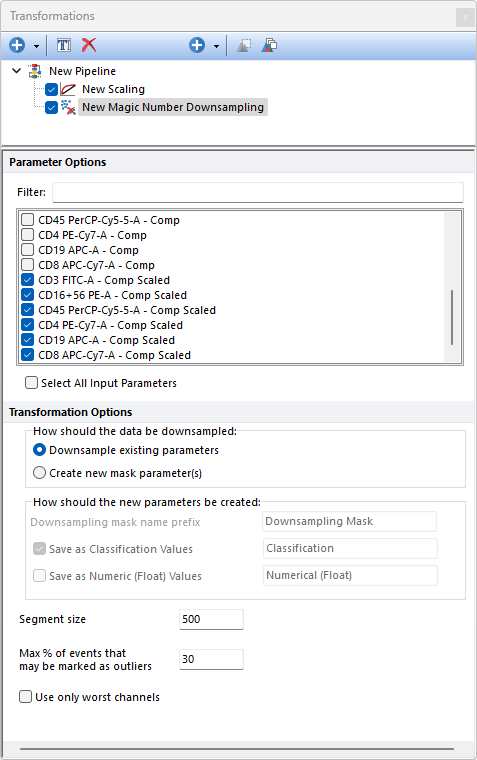
The Magic Number Downsampling pipeline step allows the user to perform quality control on flow cytometry data in order to improve both manual and automated downstream analysis. The quality control is performed by default on all of the selected input parameters (please see the Use only worst channels option for variation on this behavior).
Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The Magic Number Downsampling pipeline step separates events along the time axis into equally sized segments (the minimum number of segments is 3; the number of events in each segment is defaulted to 500 but can be customized through the Segment size field; see below) and calculates if any of the segments should be removed because they are statistically different than the others.
The cleaning is performed only when any of the below thresholds are reached:
1.MaxOfMeans: The maximum range of the means of each segment over all channels. For each channel, the 2nd and 98th percentile intensities are calculated. After calculating the mean intensity for every segment, the difference between the maximum and the minimum segment means in that channel is calculated. This is the maximum intensity drift in that channel. If this drift is larger than 15% of the difference between the 2nd and the 98th percentile intensity in that channel (i.e the MaxOfMeans threshold), then the file has to be cleaned. The MaxOfMeans value is hard-coded to 0.15 to mimic the original FlowCut algorithm. 2.MeanOfMeans: The average range of the means of each segment over all channels. The maximum intensity drift is calculated for all channels and then averaged. If the average is larger than then the MeanOfMeans threshold, then the file needs cleaning. The MeanOfMeans value is hard-coded to 0.13 to mimic the original FlowCut algorithm.
MeanOfMeans and MaxOfMeans thresholds essentially set a restriction for a tolerable mean drift.
3. MaxContin: The changes of the means between adjacent segments in each channel. The parameter help locate abrupt mean changes such as spikes. If adjacent segments have differences in their means that exceed MaxContin, then the file will undergo cleaning. The MaxContin value is hard-coded to 0.1 to mimic the original FlowCut algorithm.
If any of the above quality controls are not passed, the dataset undergoes the cleaning procedure as follows:
Eight measures of each segment (mean, median, 5th, 20th, 80th and 95th percentile, second moment (variance) and third moment (skewness)) are calculated. For each segment, all said measurements are summed all together for all channels resulting in each segment being represented by only one statistical number (i.e. the sum of all measurements in all channels). The density of summed measures distribution is then calculated. The distribution, accompanied with two hard-coded parameters (MaxValleyHgt and MaxPercCut), will determine which segments are significantly different from the other segments. The segments that are significantly different will have all of their events removed.
The Magic Number Downsampling pipeline step runs by default on all selected input parameters. However, sometimes abnormal behaviors are only present in a few channels. The outlier events in these channels can be washed out when taking into consideration all measures in all channels. As a result, outliers in these channels are not properly removed. In these cases, FCS Express can use the Use only worst channels option to only focus on these specific channels.
A limit to the number of removed events can be set through the Max % of events that may be marked as outliers setting.
The How should the data be downsampled option allows two choices: oDownsampling Existing Parameters. No mask parameter is created and only the downsampled events will be available downstream this pipeline step. oCreate new mask parameters(s). One (or two; see below) new parameter is created. The name and the nature of this parameter can be customized with the How should the new parameter be created option (see below).
The How should the new parameters be created option allows to: oDefine the Downsampling mask names prefix. This is defaulted to Downsampling Mask but can be edited. Editing this prefix is especially useful to distinguish mask parameters generated by different downsampling pipeline steps. oDefine whether the new parameter should contain data saved as Classification Values. When this option is used, sampled events will be assigned with a "Include" value in that parameter while unsampled events will be assigned with a "Exclude" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter. oDefine whether the new parameter should contain data saved as Numeric (Float) Values. When this option is used, sampled events will be assigned with a "1" value in that parameter while unsampled events will be assigned with a "0" value in that parameter. The editable field at the right of this option allows to customize the suffix to use for this parameter.
Note: when both the Save as Classification Values and the Save as Numeric (Float) value options are used, two mask parameters will be created.
|
|