Miscellaneous
The table below describes the Miscellaneous pipeline steps that are available in FCS Express. If you would like to recommend additional steps to be provided with FCS Express, please contact support@denovosoftware.com.
|
|||
|---|---|---|---|
Step
|
Description
|
||
|
The Classifications Folder allows the user to run pipeline steps for each data file that is part of a merged data file, independently, based on the classifications contained in a classification parameter. Classification parameters may be derived from file merging, downsampling masks, clustering and other pipeline steps.
Although any pipeline step can be run within the Classifications Folder, it is effectively used to run data pre-processing steps like Scaling, Normalization, 0-1 Scaling, Downsampling, FlowAI, and FlowCut on each data file contained within a larger merged data file.
As an example, in the pipeline below (right) a Classification Folder has been used to run three pre-processing steps, Scaling, Gating and Downsampling, based on the classification parameter of a merged file (left). Note that subsequent steps, such as UMAP and tSNE have been placed downstream of the classification folder step so those steps will be run after pre-processing and on the merged data. In the example below, Scaling, Gate Downsampling, and Interval Downsampling, will be run for each of the three donor samples, the resulting pre-processed data will then be run as an entire merged file for uMAP and tSNE.
|
||
|
The Folder element allows users to add a folder within the pipeline to better organize and group steps in the pipeline. The Pipeline Folder has a check-box that allows users to easily activate/deactivate all the steps contained in the Pipeline Folder.
To rename a Pipeline Folder, select the folder by left clicking on it and the do either of the following: •Click on the •Press F2 on the keyboard; •Right click on the folder and select Rename.
|
||
|
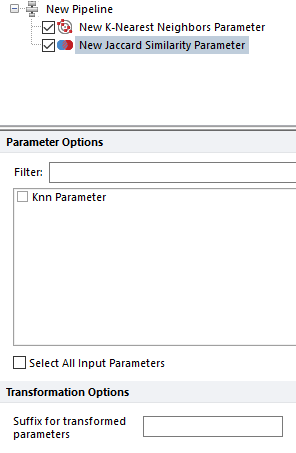
The Jaccard Similarity Parameter step calculates the Jaccard Index (https://en.wikipedia.org/wiki/Jaccard_index) for every pair of events provided as the input data. The Jaccard index is a measure of overlap between two sets, such that the Jaccard index equals one if the sets are identical and zero if the sets do not contain common members. This step can be used to calculate the degree of overlap between sets of event attributes. The input is a two-dimensional array of event attributes. This step should be preceded in the Pipeline by a K-Nearest Neighbor Parameter step (see next entry in table). The output is a square two-dimensional array of Jaccard index scores, J.
|
||
|
This pipeline step allow to define the nearest neighbors for each the input data point. The neighbors are calculated in the space defined by the parameters selected in the Parameter Options list (see image below).
Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The number of nearest neighbors to output is defined by the user through the Number of Neighbors field. The output of this pipeline step is a new parameter in which nearest neighbors are listed for each input data. Said new parameter can be visualized through the Data Grid.
|
||
|
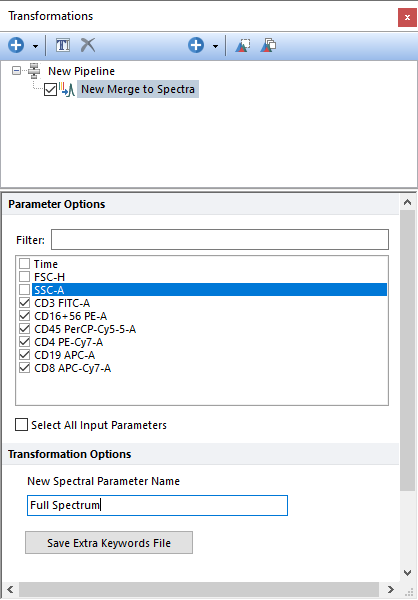
The Merge to Spectra step allows users to create a virtual spectrum parameter by combining multiple input detectors, generally from an overdetermined system. The derived spectrum parameter can then be displayed using a Spectrum Plot and results used for the Unmixing pipeline step. Please see our chapter on Virtual Spectrum for more information.
Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
The virtual spectrum parameter will be named with the text defined in the New Spectral Parameter Name field. In the example below, the new parameter has been named Full Spectrum.
The Merged Spectra details can also be saved as an AllExtraKeywords.txt file and then be used to automatically recreate the spectrum parameter without using any pipeline. Please refer to the Virtual Spectrum chapter for more details.
|
||
|
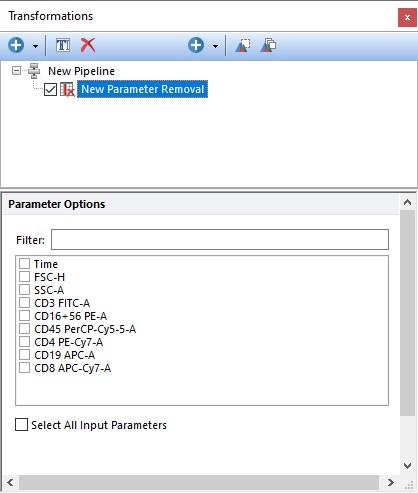
The Parameter Removal step allows users to remove parameters from the parameter list. Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
Parameter Removal is particularly useful when one or multiple parameters are not useful in the analysis, or a data file for use in a template was acquired with more parameters than the original data file, and thus can be removed to make the parameter list shorter or to make the data file adhere to the original template design.
|
||
Python Transformation
|
Please refer to the Python Transformation chapter for more details. |
||
|
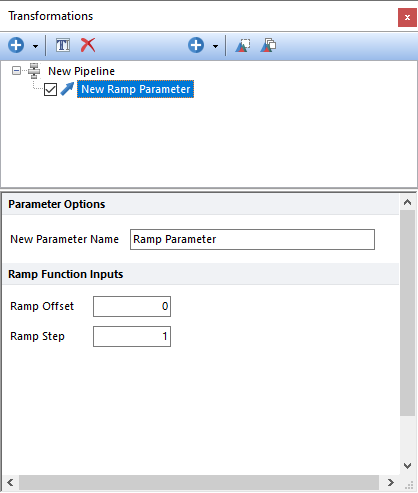
The Ramp Parameter step creates a new parameter when ever events are assigned with an increasing integer number. The Ramp Offset field allows users to define the starting value, while the Ramp Step filed allows users to define the step between each value. If the Ramp Offset and the Ramp Step are set to 0 and 1 respectively, and the pipeline is applied to a population that counts 100 events, the Ramp Parameter will number events from 0 to 99.
|
||
|
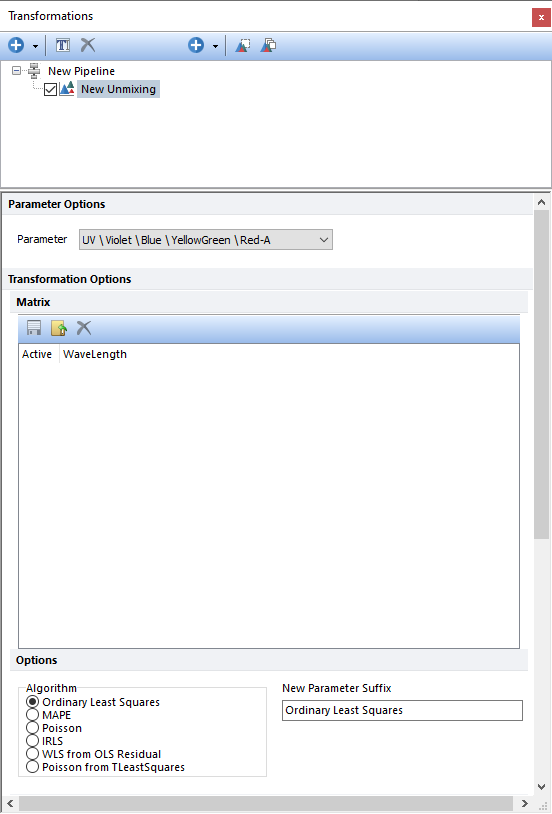
The Unmixing step allows a user to perform spectral unmixing on the input spectral parameter using an unmixing matrix imported by the user.
The spectrum parameter may come from a data file acquired on a compatible spectral instrument or from a virtual spectrum parameter created via the Merge to Spectra step. The unmixing matrix can also be one that was created in FCS Express using the Unmixing wizard.
In the Options section, the user can define: •Which algorithm use to unmix the input parameter. •Which suffix to add to the unmixed parameters.
|
||
|
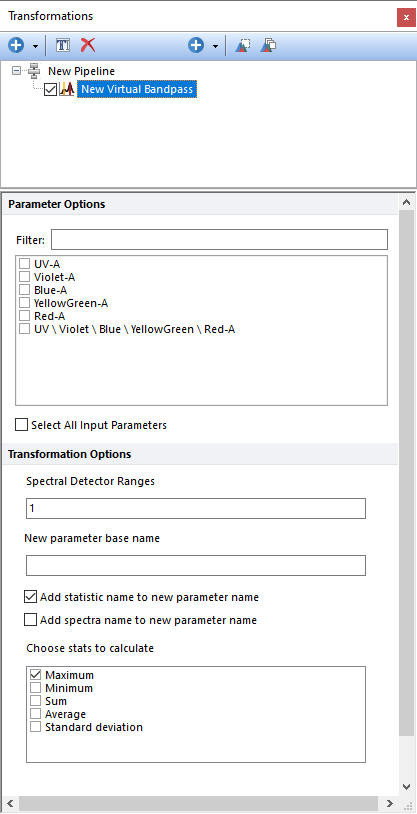
The Virtual Bandpass step allows users to combine multiple single detectors, generally from an overdetermined system (e.g. a spectral instrument) to recreate a virtual bandpass filter including a particular range of detectors. Parameters can be filtered or sorted to assist in the selection of parameters when multiple parameters are available in the template file. Using the Filter: field a user can remove unwanted parameters from view to simplify selection. For example, if a user only wanted to select area parameters typing "-A" in the field would reduce the number of parameters seen in the parameter list. By right clicking in the parameters section, you can use Sort Ascending, Sort Descending, or Unsorted to easily manage parameter ordering and facilitate parameter selection. In the right click menu, you can easily select parameters by utilizing Check All, Uncheck All, Check Selected, Uncheck Selected, Invert Selection on All. The options Check Selected and Uncheck Selected allow for using the shift key or Ctrl key to multi-select parameters and check or uncheck them all simultaneously.
Compatible input parameters for the Virtual Bandpass step are spectral parameters that are derived from a compatible spectral data file or from a Merge to Spectra pipeline step. If multiple spectral parameter are selected, the virtual band pass filter will be applied to each of them independently.
Once the input parameters are defined, the user can specify which detectors to combine in the Spectral Detectors Range field. Detectors may be listed as comma separated values (e.g. 1,2,3,4,5) or with dash (e.g. 1-5 is equal to 1,2,3,4,5). Detector numbers are associated with the order in which they appear in a spectral plot. For instance, if your spectral plot displays FITC-A, PE-A, PeCy7-A, APC-A, and APC-Cy7-A, the detector numbers will be 1. FITC-A, 2. PE-A, 3. PeCy7-A, 4. APC-A, and 5. APC-Cy7-A (example below).
The calculated values for each Virtual Bandpass Filter step will be saved into a new parameter. The new parameter is defined by the user in the New parameter base name field (e.g. "UV-A Virtual bandpass").
Different summary statistics (i.e.: Maximum, Minimum, Average, Average, Standard Deviation) can be selected for the calculation of the new values.
Two suffix can be automatically added to the new parameter name via the following check boxes: •Add stats name to new parameter base name. When this check box is checked, the name of the statistics selected from the Chose stats to calculate list will be added as a suffix to the name defined in the New parameter base name field. If multiple statistics are selected, the name of the statistics will always be added as a suffix. •Add spectra name to new parameter base name. When this check box is checked, the name of the parameter selected from the Parameter Options list will be added as a suffix to the name defined in the New parameter base name field. If multiple input parameters are selected, the name of input parameter will always be added as a suffix.
|
||