Working with k-means using 1D-2D plots
When a k-means transformation includes parameters other than Cluster Assignments, and these are applied to 1D or 2D plot, the clustering parameters can be accessed as standard flow cytometry parameters.
In this tutorial we will access clustering parameters on 1D and 2D plots, and we will use gates and markers to select clustered events. These gates and markers will then be available for downstream analysis.
For this tutorial we will use the k-means on plots.fey layout stored in the k-means tutorial subfolder of the Tutorial Sample Data folder.
To open the layout required for this tutorial:
1.Select the File tab→Open.
2.Navigate to the Tutorial Sample Data folder and open the k-means on plots.fey layout stored in the k-means tutorial subfolder.
The layout should appear as shown in Figure T33.10:
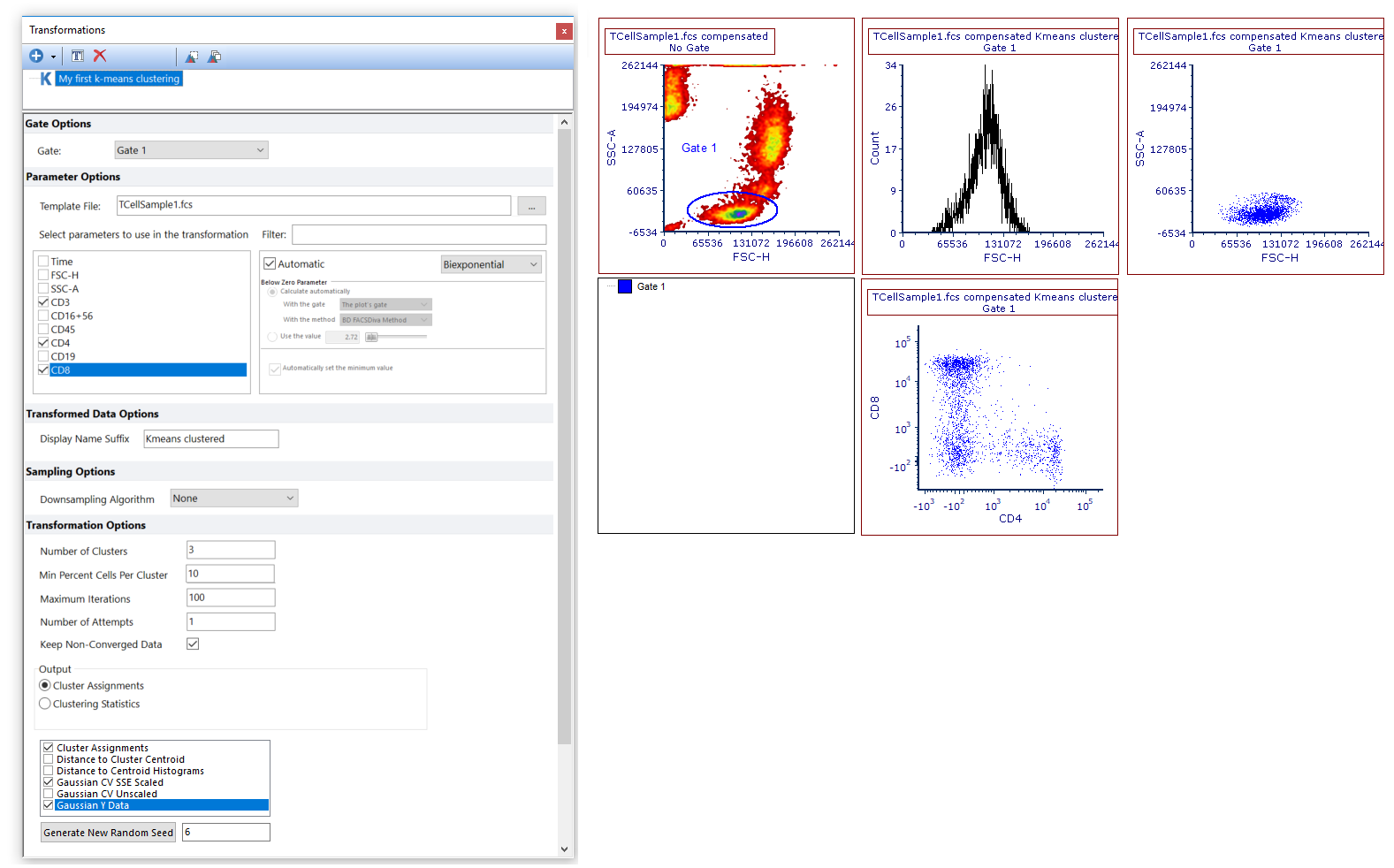
Figure T33.10. The k-means on plots.fey layout as it appear after opening it.
In this layout a k-means transformation has been created and applied to a histogram and to 2 color dot plots, (please note the Kmeans clustered suffix in the plot title). In order to simplify gate visualization, and to create a cluster gate with the correct hierarchy, Gate 1 (thus the gate on which the clustering has been performed), has already been applied to all this objects.
In this example, the following clustering parameters have been selected, and thus automatically created as output results for the k-means clustering: Cluster Assignments (which is suitable for Heat Maps, see the next section of this tutorial), Gaussian CV SSE Scaled and Gaussian Y Data (which can be both accessed on 1D and 2D plots). To learn more about the available clustering parameters, please refer to the Defining a k-means cluster analysis topic of the manual.
We will now select clustered events using markers on 1D plots.
3. Change the parameter displayed on the 1D plot to be Kmeans 3 Gaussian CV SSE Scaled. Three peaks will appear in the 1D plot, one for each clusters (Figure T33.11).
4. Select the peak on the left (i.e. cluster 1) by creating a marker.
5. Convert the marker to a gate, by right clicking on it and selecting Convert Marker to gate → Marker #1 (M1) → Convert and Link. A Create a new gate dialogue opens.
6. Replace Gate 2 with Marker on Cluster as gate name and click OK.
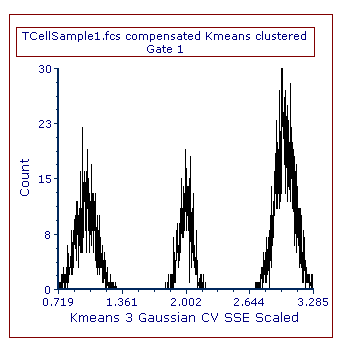
Figure T33.11. A 1D plot displaying a clusering parameter.
The content of the layout should now be similar to Figure T33.12. Please note that the new gate is now listed in the gate view, and that the events belonging to Marker on Cluster are now shown in green in both of the color dot plots.
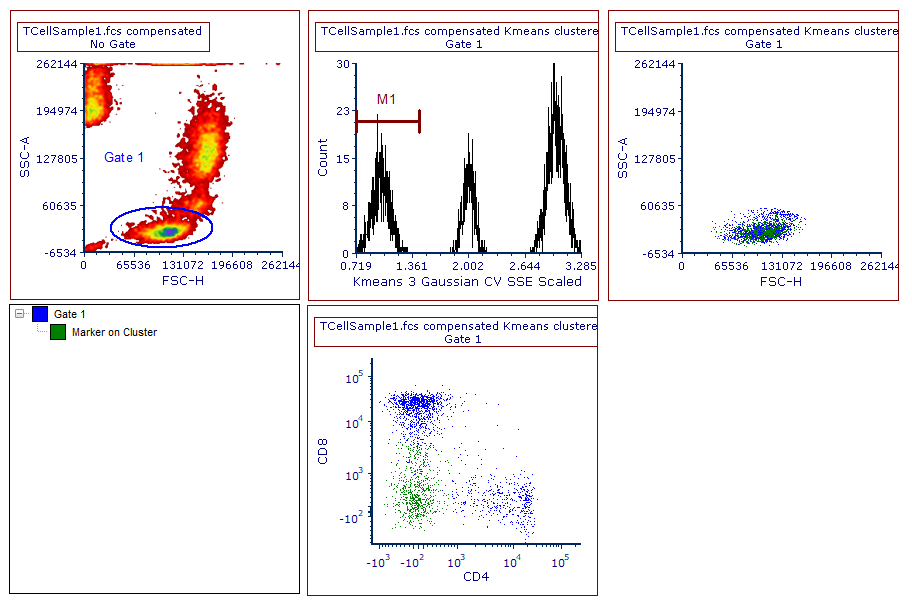
Figure T33.12. A marker has been created to select cluster 1. The marker has then been converted and linked to be a gate. The gate view and the color dot plots reflect this change.
7. Move marker M1 to select a different peak in the histogram, thus a different cluster. Please take note that the color dot plot is updated based on these changes.
Also, please note: Since the M1 marker has been converted to a gate, now it can also be applied to a plot and to other objects in the layout. The M1 gate will have the same functionality as any other gate.
We will now select clustered events using gates on 2D plots.
8. Change the parameter displayed on the 2D color dot plot in the upper part of the layout to be Kmeans 3 Gaussian CV SSE Scaled on the X axis, and Kmeans 3 Gaussian Y Data on the Y axis (Please also note that clustering parameters can be used together with any other non-clustering parameter as well on 2D plots).
The content of the plot will be similar to the one displayed in Figure T33.13. Please note that the coloring may be different depending on the position of marker M1 (see step 7).
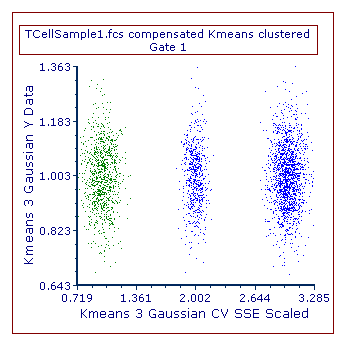
Figure T 33.13. A 2D plot displaying two clustering parameters.
9. Select one of the blue clusters by creating a 1D Gate (select Gating→Create Gates→Markers from the ribbon bar) around it. A Create New Gate dialogue opens.
Note: although any type of gate can be used to select clustered events on 2D plots, 1D Gates are particularly useful in this cases as they automatically extend from the bottom to the top of the 2D plot.
10. Replace Gate 2 with Gate on Cluster as gate name, change color to be red and click OK.
The layout should now appear similar to the one displayed in Figure T33.14. Please note that the new gate (i.e Gate on Cluster) is now listed in the gate view, and that the events belonging to it are now red, in both of the color dot plots.
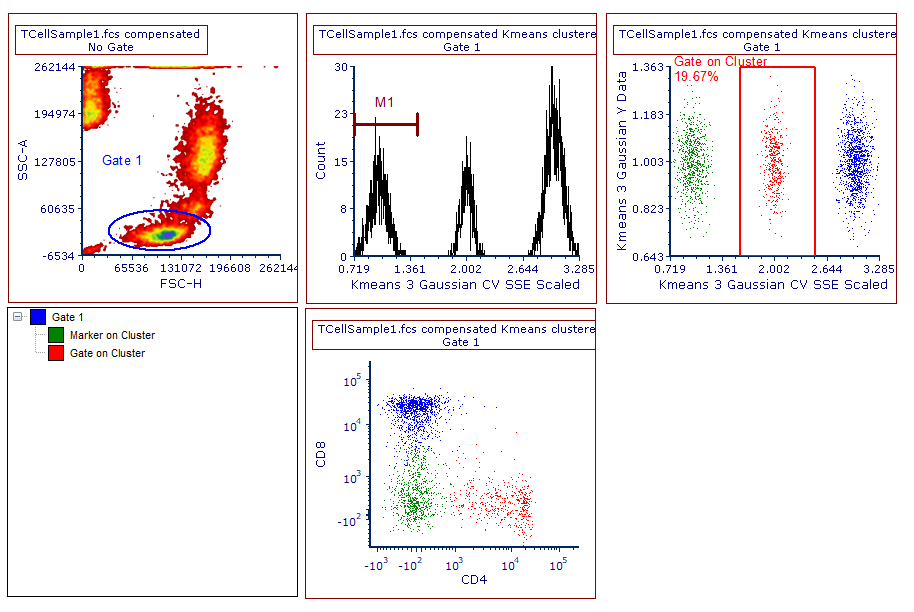
Figure T33.14. A gate has been created to select cluster 2. The gate view and the color dot plots reflect this change.
11. Move the Gate on Cluster gate to select a different cluster. Note that the color dot plots update their content based on these changes. Note: the Gate on Cluster gate can also be applied to a plot and to other objects in the layout. The Gate on Cluster gate will have the same functionality as any other gate.
